
The first evidence of in vivo SARS-CoV-2 escape from antibodies: emergent Spike deletion H69/V70 and D796H mutation in a convalescent plasma (CP) treated patient. These mutations conferred reduced susceptibility to the CP and sera from multiple donors. medrxiv.org/content/10.110…
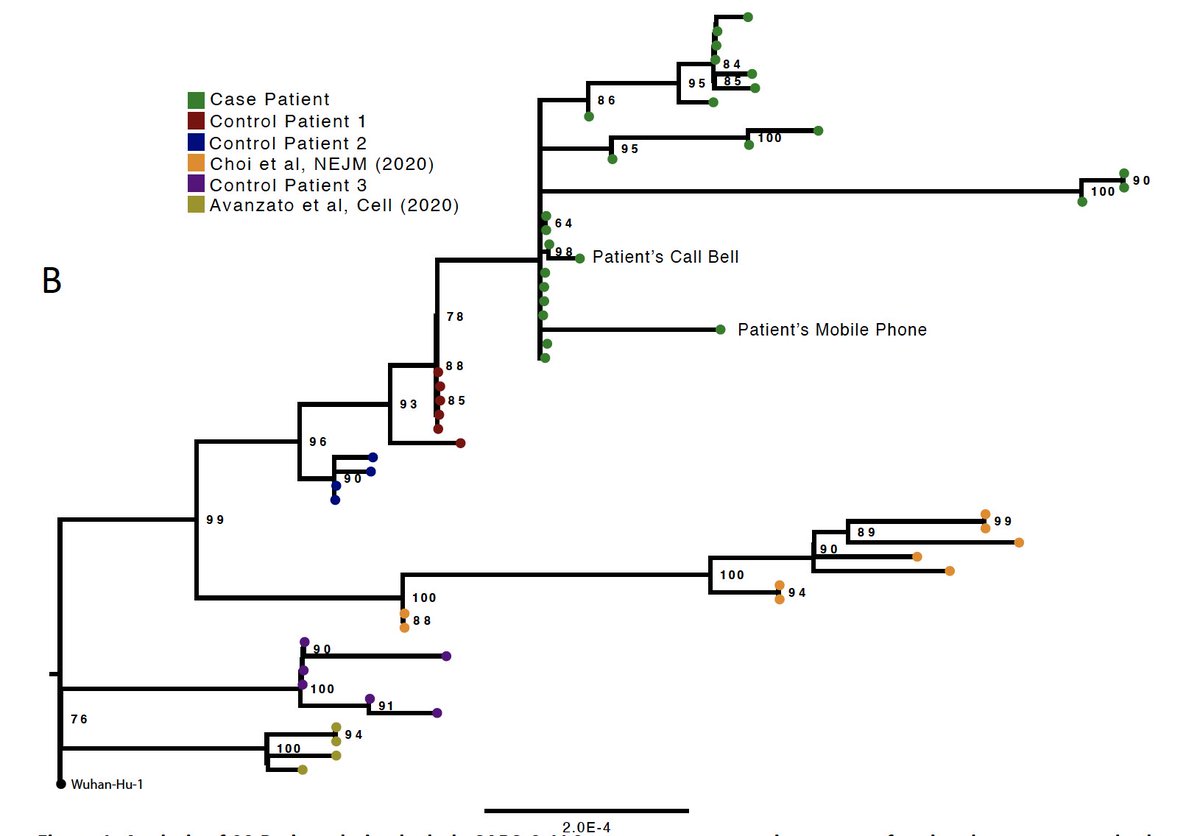
Phylogenetic analysis of published cases demonstrates diversity of SARS-COV-2 virus in long term shedders. This case is in green. 


Dramatic viral population structure change after receiving 2 units of CP at days 63 and 65 with the emergence of ΔH69/V70 and D796H mutants at frequencies of 90%. Following CP washout a population bearing Y200H
and T240I emerged and then a highly divergent

and T240I emerged and then a highly divergent

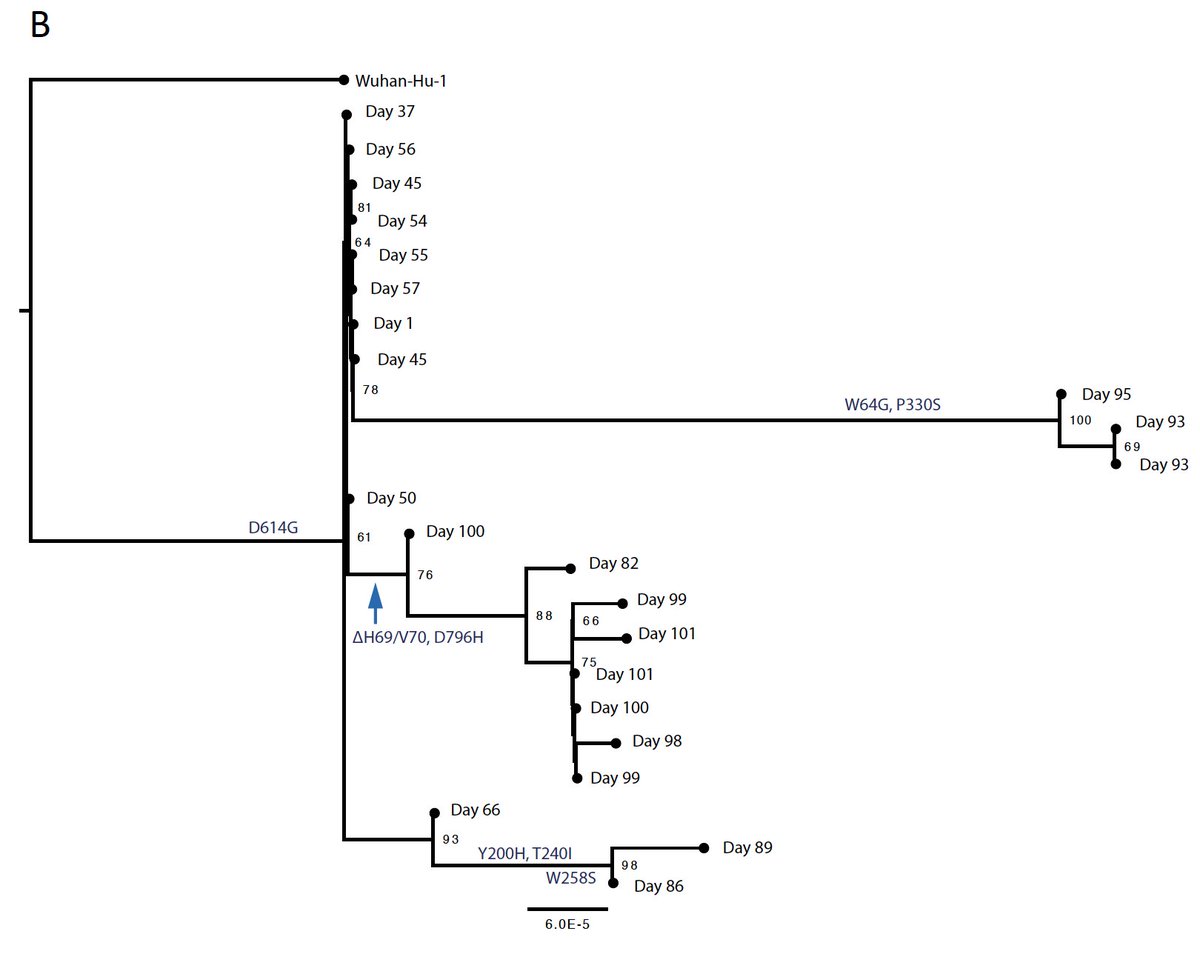
population bearing W64G and P330S mutations in Spike at day 93. Following a 3rd course of CP on day 95, the ΔH69/V70 and D796H virus population re-emerged to become the dominant viral strain reaching variant frequencies of >90%.
We used Spike pseudotyped lentivirus to test neutralisation of the WT and ΔH69/V70+D796H mutant by the administered CPs and the patient sera on days following the CP infusions. We found that the mutant was less sensitive to antibody neutralisation. 
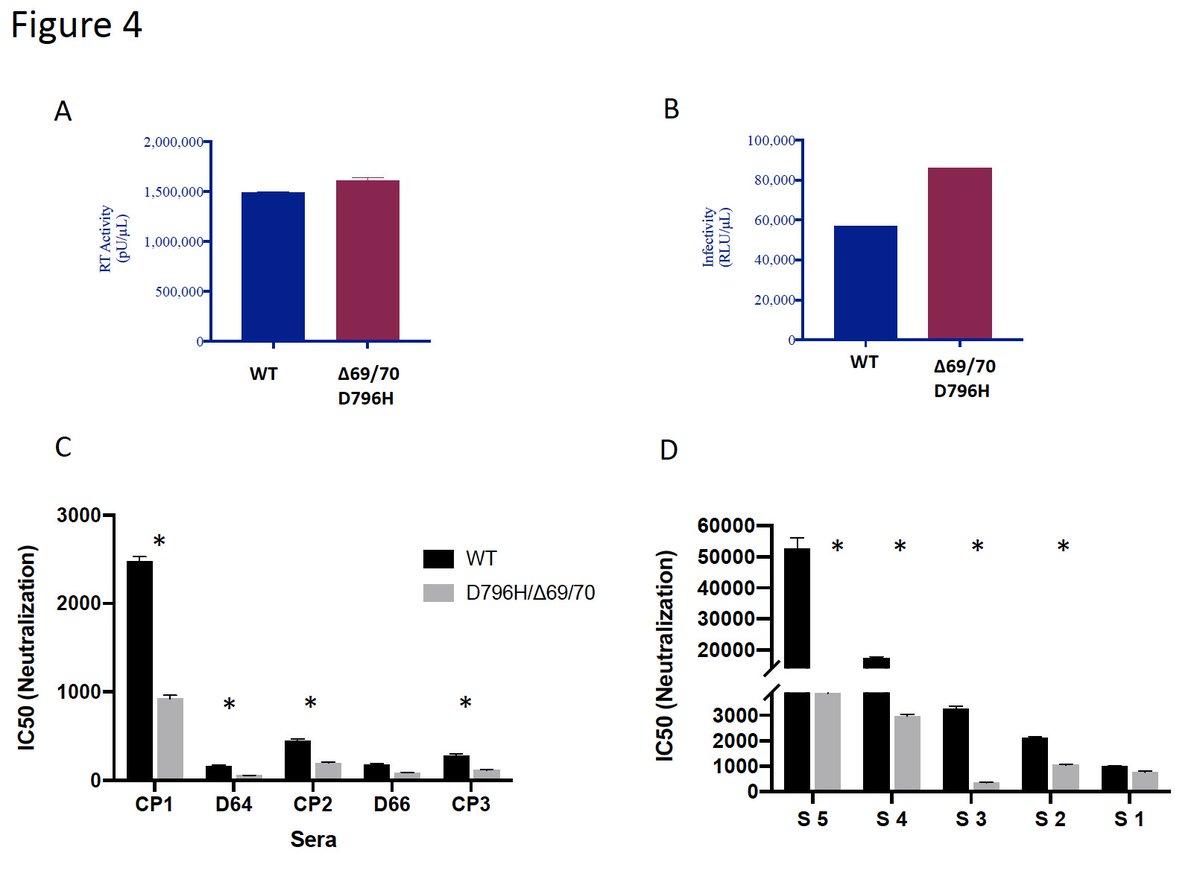
We also tested neutralisation by sera from recovered patients with confirmed COVID-19 and also found that the combined ΔH69/V70 and D796H mutant was less sensitive to neutralisation. These observations suggest that theses mutations represent a broad escape mechanism.
ΔH69/V70 and D796H are in the S1 NTD and S2 respectively. They are both in exposed, poorly resolved loops which may represent antibody epitopes. We hypothesise that mutations at these sites may reduce antibody binding and efficacy. 

• • •
Missing some Tweet in this thread? You can try to
force a refresh



