
Introducing IN SITU GENOME SEQUENCING, a new technology to resolve DNA sequence🧬 and structure🔬
A team effort with @Andrew_C_Payne, @PaulReginato, @eboyden3, @JD_Buenrostro, and Fei Chen
science.sciencemag.org/content/early/…
This is how it works: (1/8)
A team effort with @Andrew_C_Payne, @PaulReginato, @eboyden3, @JD_Buenrostro, and Fei Chen
science.sciencemag.org/content/early/…
This is how it works: (1/8)
We started with a basic question: how can we sequence DNA while preserving 3D genome structure?
Instead of extracting DNA, we developed a method to construct sequencing libraries within fixed nuclei (2/8)
Instead of extracting DNA, we developed a method to construct sequencing libraries within fixed nuclei (2/8)
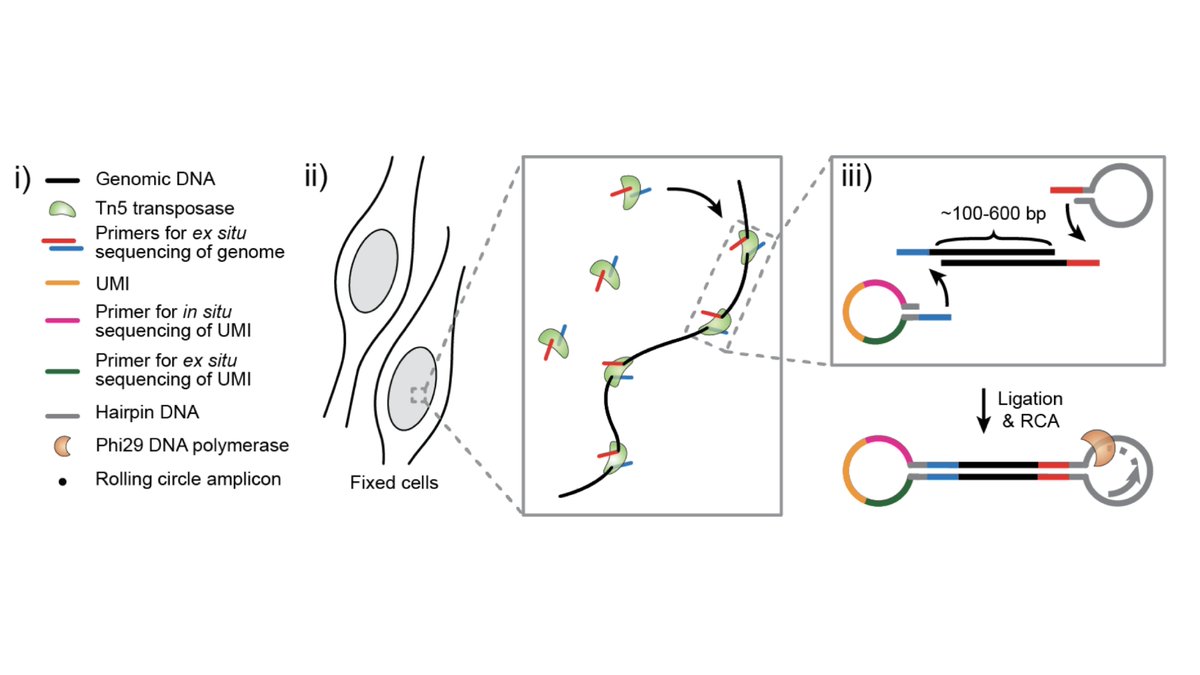
Next, we use in situ sequencing to read out the libraries via fluorescence microscopy.
This is a single nucleus: each spot is an amplified DNA fragment, the colors indicate the 4 DNA bases, and each frame represents a round of sequencing (3/8)
This is a single nucleus: each spot is an amplified DNA fragment, the colors indicate the 4 DNA bases, and each frame represents a round of sequencing (3/8)
The trick is that instead of directly in situ sequencing the genomic DNA, we read out unique spatial barcodes.
Then we perform Illumina sequencing and integrate across barcodes, yielding 100-1000s of spatially-resolved reads per nucleus (4/8)
Then we perform Illumina sequencing and integrate across barcodes, yielding 100-1000s of spatially-resolved reads per nucleus (4/8)
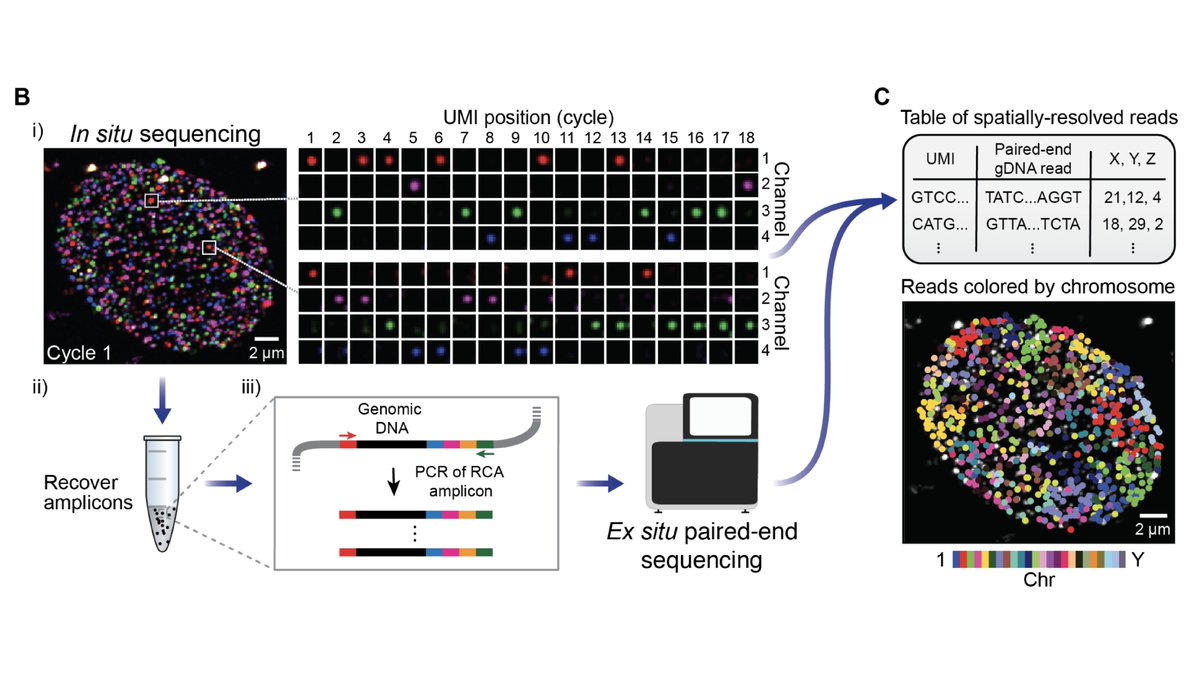
We first examined known features of spatial genome organization in cultured PGP1 (aka @geochurch) fibroblasts.
We then applied our method to intact early mouse embryos at the zygote, 2-cell, and 4-cell stage (5/8)
We then applied our method to intact early mouse embryos at the zygote, 2-cell, and 4-cell stage (5/8)
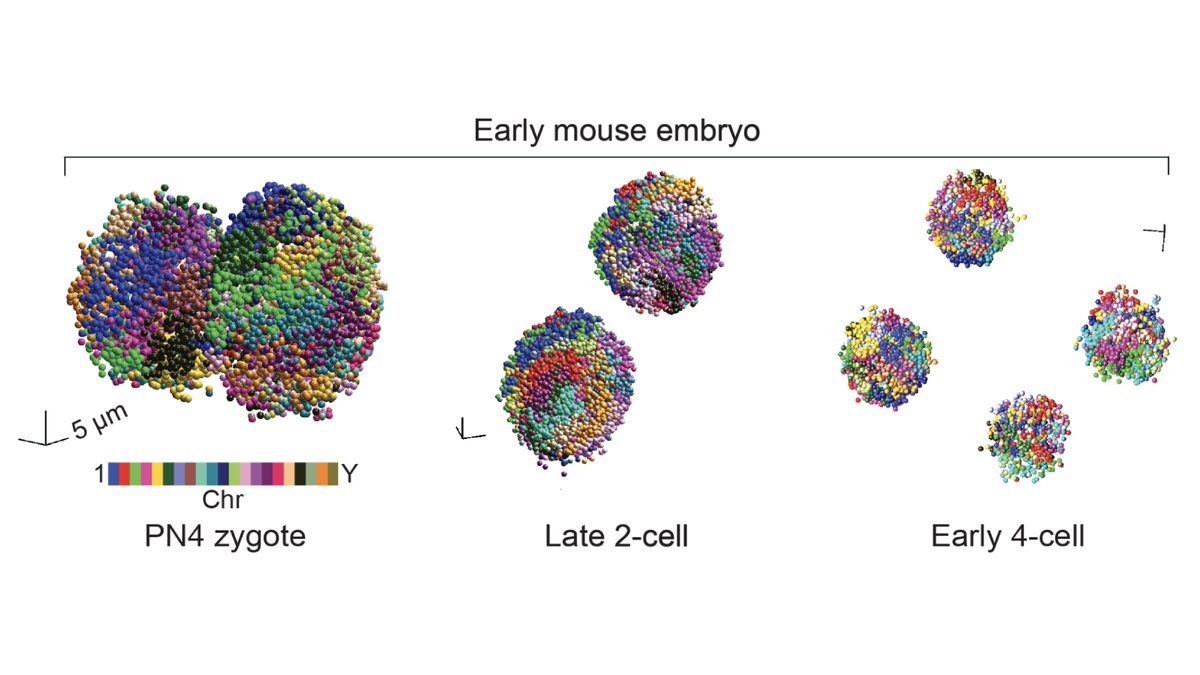
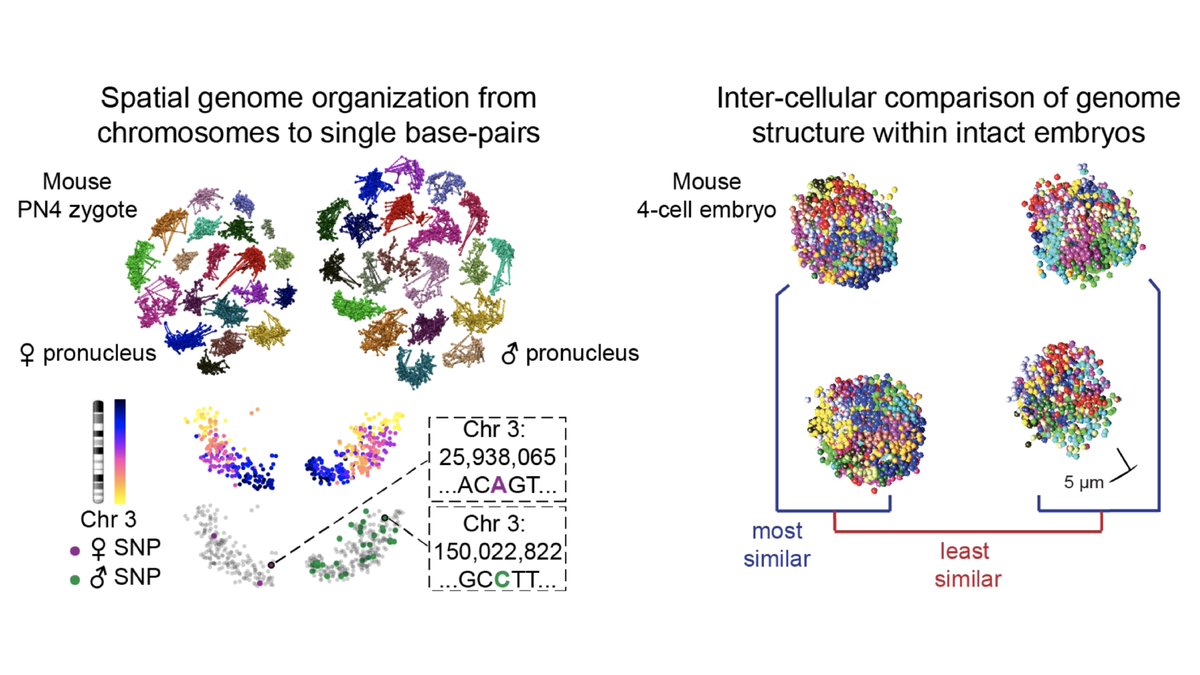
Our method is sequencing-based, so we can leverage genotype information to compare maternal and paternal chromosome organization across developmental stages.
Our method is also imaging-based, so we can compare structures across cells within a single intact embryo (6/8)
Our method is also imaging-based, so we can compare structures across cells within a single intact embryo (6/8)
Check out the paper for more! Thanks to all co-authors, reviewers, and the editorial team @ScienceMagazine who helped us sneak this in at the buzzer of this very weird year (7/8)
science.sciencemag.org/content/early/…
science.sciencemag.org/content/early/…
If you'd like to explore the data for yourself, head on over to our interactive Shiny app! (8/8)
buenrostrolab.shinyapps.io/insituseq/
buenrostrolab.shinyapps.io/insituseq/
• • •
Missing some Tweet in this thread? You can try to
force a refresh


