
I’m super excited about our new paper in @EUplatinum, “Nascent Prostate Cancer Heterogeneity Drives Evolution and Resistance to Intense Hormonal Therapy” in @sowalsky lab @NIH @NCIResearchCtr @PCFScience 

Patients diagnosed w/ aggressive, non-metastatic prostate cancer are at higher risk of recurrence after radical prostatectomy (RP). Post-op, some tumors progress to castration-resistant prostate cancer. Are there tumors that will respond to pre-surgical (neoadjuvant) therapies?
A trial recently completed at @NCIResearchCtr clinical trial enrolled patients with high risk, localized prostate cancer, who had 6 months of intense neoadjuvant ADT followed by RP (clincancerres.aacrjournals.org/content/27/2/4…). We found that 40% of patients responded to treatment. 

We first asked whether we could identify from biopsies acquired prior to treatment genomic alterations that distinguished exceptional responders (ERs) from incomplete and nonresponders (INRs). Not surprisingly, TP53 and PTEN mutations were enriched in INRs. 
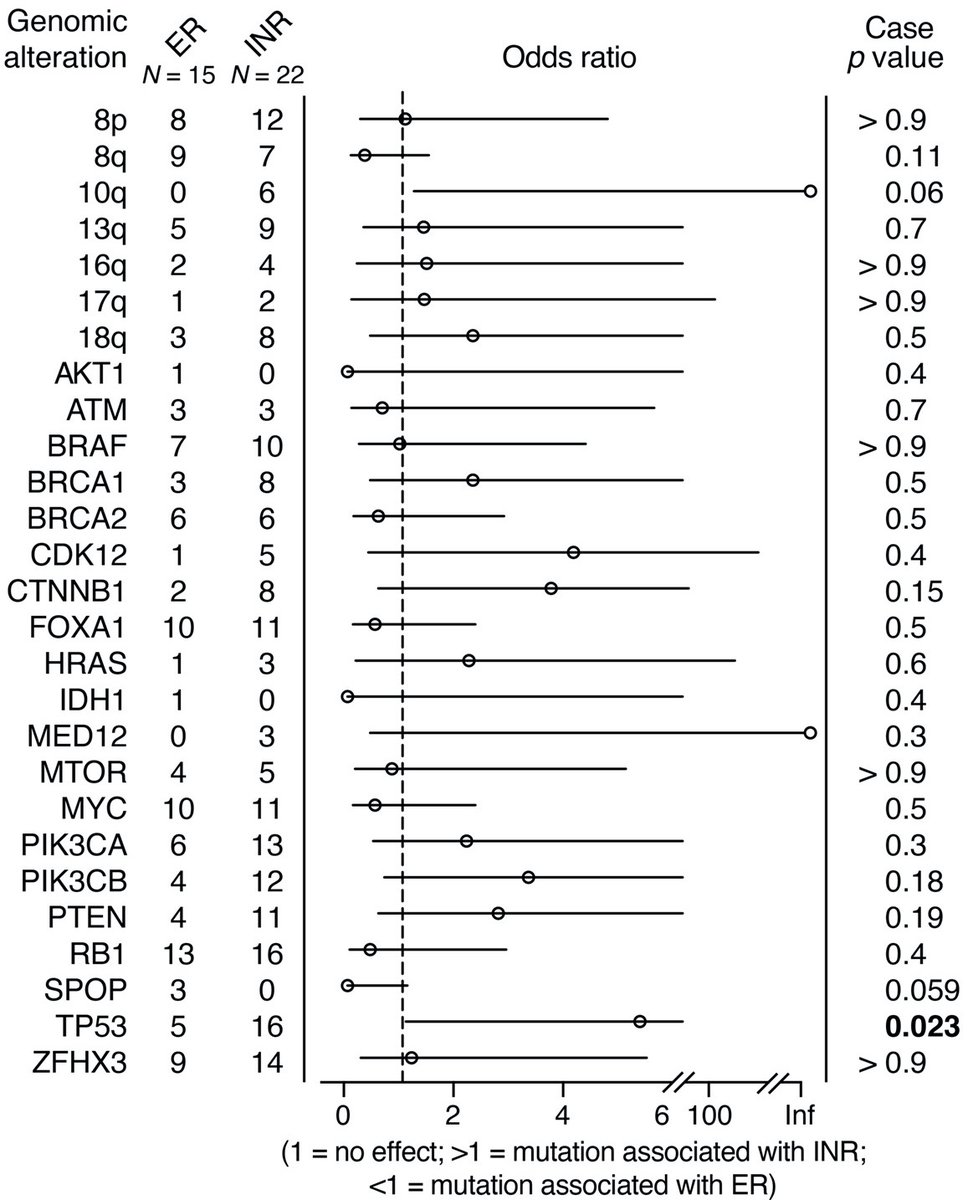
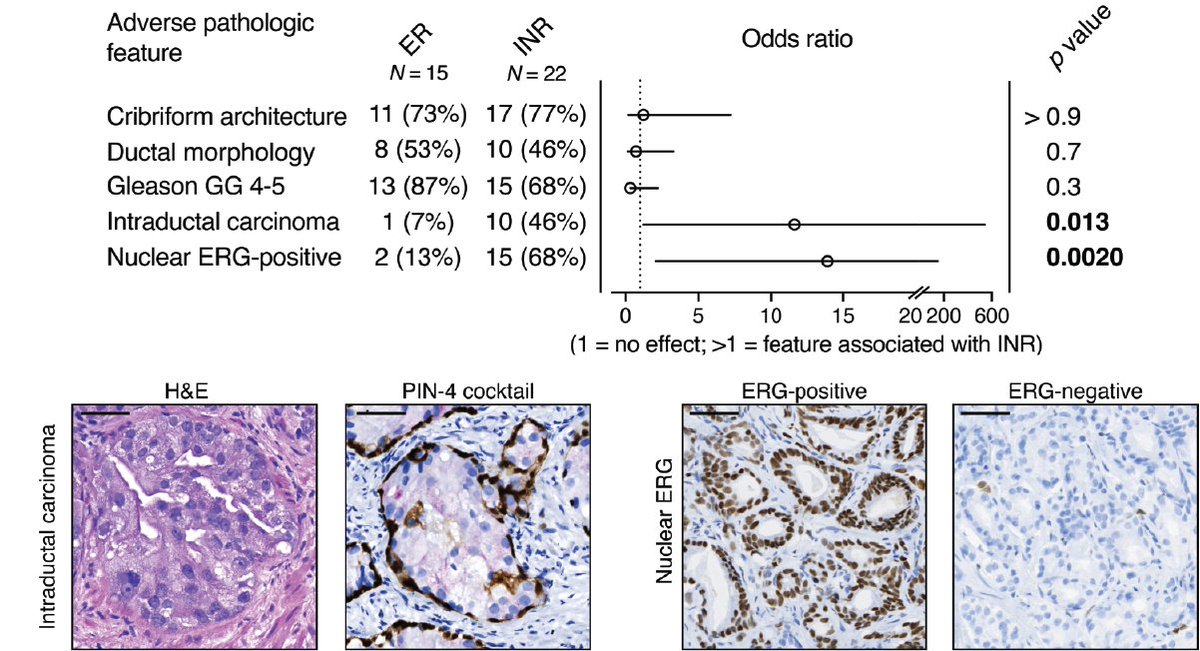
We also asked whether there were any histologic features that tracked with INRs. Notably, ERG expression and intraductal carcinoma (IDC-P) were enriched in the INRs. 
Our comprehensive analysis of over 100 tumor biopsies included a panel of prostate cancer IHC stains, including tumor cell expression of PSA. The more PSA a tumor expressed, the more likely it was going to respond to therapy. 

Because we used software to analyze our IHC, we also observed heterogeneity in per-cell stain intensities. For PSA, the less intense the stain, the more variable it was, indicating a more diverse population of tumor cells, which we calculated using the Shannon diversity index 

When we looked back the genomic data we generated from over 100 biopsies, we found that the somatic copy number profiles of multiple biopsies from the same patient were more similar to each other in ERs than in INRs. 

This made us question whether the apparent genomic diversity of a tumor was representative of multiple tumor cell populations. We used a bioinformatic approach to compile all genomic data from each patient to assemble phylogenetic trees 

We next took the top genomic and histologic features that were enriched in INRs (IDC, chr10q loss, ERG, and TP53) and used them to build a prediction model for response to intense neoadjuvant ADT. On their own, a probability cutoff of 60% predicts poor response with AUC=0.89. 

This prediction model correlated well with genomic heterogeneity. When we computed both genomic heterogeneity and a prediction of poor response using these factors from high-risk prostate TCGA cases, genomic diversity again correlated with INR probability. 

We know from previous studies that patients with very good responses to intense neoadjuvant ADT are at far less risk of recurrence. However, TCGA cases predicted to respond to therapy still recurred at the same rate, meaning there is a subset of patients who may benefit. 

By combining pre-tx imaging with our model, the prediction model improved to AUC=0.98. This simple model of only 5 features may show great promise in selecting pts most likely to benefit. If a tumor is determined to be INR, pts could be treated differently, saving valuable time. 

All of this work over the past 5 years would be impossible without the help of so many. Thank you to all the patients who participate in these studies. Thanks to @sowalsky for his mentoring. And of course, our funding sources @PCF_Science @cdmrp @IRPatNIH
A tremendous volume of data accompanies this paper, both in the Supplementary data, as well as raw data deposited in dbGaP and TCGA. #openaccess #datasharing. We look forward to seeing similar datasets deposited so we can validate our findings, and others can validate theirs
• • •
Missing some Tweet in this thread? You can try to
force a refresh



