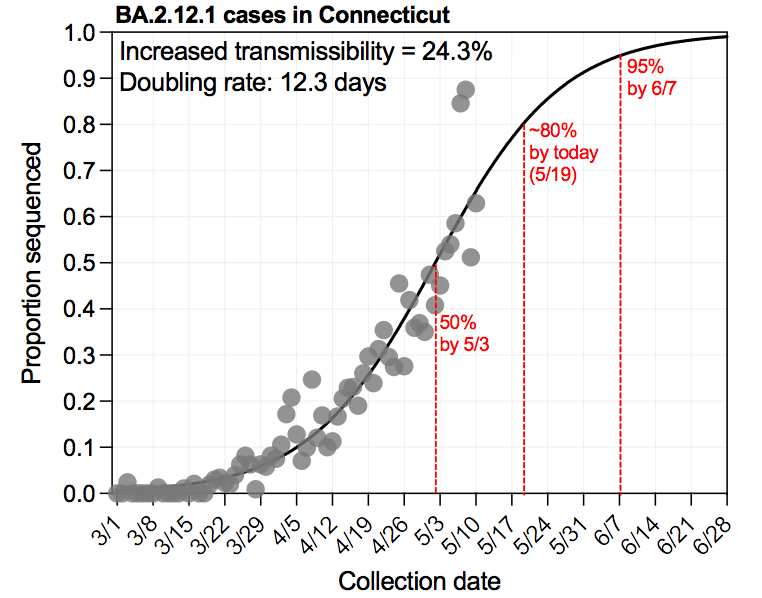
🧬4/1 Connecticut #SARSCoV2 variant surveillance report @CovidCT | @jacksonlab | @CTDPH | @YaleSPH
- B.1.1.7 *may* be leveling off
- B.1.526 continues to 📈
- More B.1.351, P.1, & E484K cases
Short 🧵 | Report 👇
covidtrackerct.com/variant-survei…
- B.1.1.7 *may* be leveling off
- B.1.526 continues to 📈
- More B.1.351, P.1, & E484K cases
Short 🧵 | Report 👇
covidtrackerct.com/variant-survei…

2/8 We didn't see an increase in the % of B.1.1.7 from the sequencing data last week, and the TaqPAth SGTF data also suggests that the expansion of B.1.1.7 *might* be slowing down in CT.
While this is good news, I think that it points to something else...
While this is good news, I think that it points to something else...

3/8 That something else is B.1.526.
It made up 32% of the sequenced cases analyzed this week, a 9% increase from the week before.
Its proportion is higher in Fairfield county, which is close to NY - where B.1.526 is currently dominating.
Data 👇 since 2/1
It made up 32% of the sequenced cases analyzed this week, a 9% increase from the week before.
Its proportion is higher in Fairfield county, which is close to NY - where B.1.526 is currently dominating.
Data 👇 since 2/1

4/8 What does the rise in B.1.526 mean?
Not quite sure yet. This lineage is complex. Some have the Spike E484K mutation (left), while others have the S477N mutation (right). We don't quite have a handle on their impact on vaccines and/or disease in the context of this lineage

Not quite sure yet. This lineage is complex. Some have the Spike E484K mutation (left), while others have the S477N mutation (right). We don't quite have a handle on their impact on vaccines and/or disease in the context of this lineage


5/8 There are now at least 8 lineages in CT with the E484K mutation, which was found on ~20% of the samples sequenced last week.
Top to bottom order on tree 👇:
- B.1.526
- B.1.314
- B.1.351
- B.1.525
- B.1.1.7
- P.1
- R.1
- R.2
Top to bottom order on tree 👇:
- B.1.526
- B.1.314
- B.1.351
- B.1.525
- B.1.1.7
- P.1
- R.1
- R.2

6/8 Even though the cases are low, we are starting to see evidence for local transmission of both B.1.351 (left) and P.1 (right) in CT.
Not sure how much room is for these variants to take hold with the growing dominance of B.1.1.7 and B.1.526. Will be very important to watch.

Not sure how much room is for these variants to take hold with the growing dominance of B.1.1.7 and B.1.526. Will be very important to watch.


7/8 This week I'd like to highlight the work by @IsabelOtt and @mallerybreban - our amazing sequencing duo! They sequence 188 samples per week, and generating fantastic data. I mean, it doesn't get a whole lot better than this 😍
COVIDseq + NovaSeq = win
COVIDseq + NovaSeq = win

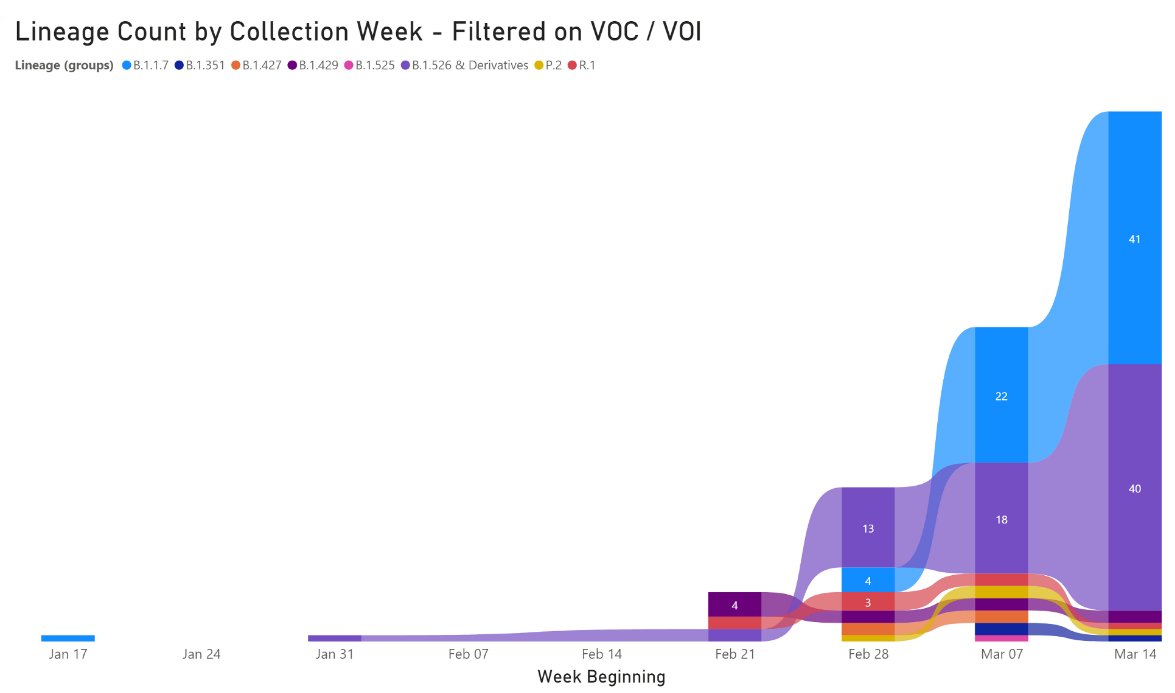
8/8 I'd also like to highlight the work by our partners @jacksonlab - led by Mark Adams 👏
They've been increasing their random sequencing and are also detecting the rapid expansion of B.1.526 (purple).
You can learn more about their work here 👇: jax.org/coronavirus-in…
They've been increasing their random sequencing and are also detecting the rapid expansion of B.1.526 (purple).
You can learn more about their work here 👇: jax.org/coronavirus-in…
• • •
Missing some Tweet in this thread? You can try to
force a refresh