
Associate Professor @YaleEMD / @YaleSPH. Studies 🦟🦠 transmission, evolution, and emergence. PI of @covidCT 🧬. Will sequence for 🍺.
2 subscribers
How to get URL link on X (Twitter) App


 It's based on a design led by @Scalene & @pathogenomenick originally for Zika virus that was adapted for SARS-CoV-2 ("ARTIC protocol") and used by labs around the world.
It's based on a design led by @Scalene & @pathogenomenick originally for Zika virus that was adapted for SARS-CoV-2 ("ARTIC protocol") and used by labs around the world. 
 Using a logistic regression of the daily frequencies, we predict that as of today (July-14), BA.5 is probably 80-90% in Connecticut.
Using a logistic regression of the daily frequencies, we predict that as of today (July-14), BA.5 is probably 80-90% in Connecticut. 

 Omicron BA.2.12.1 is still 📈 in Connecticut as it is across most of the US. Fitting the % of sequenced cases to a logistic growth curve, we estimate that BA.2.12.1:
Omicron BA.2.12.1 is still 📈 in Connecticut as it is across most of the US. Fitting the % of sequenced cases to a logistic growth curve, we estimate that BA.2.12.1: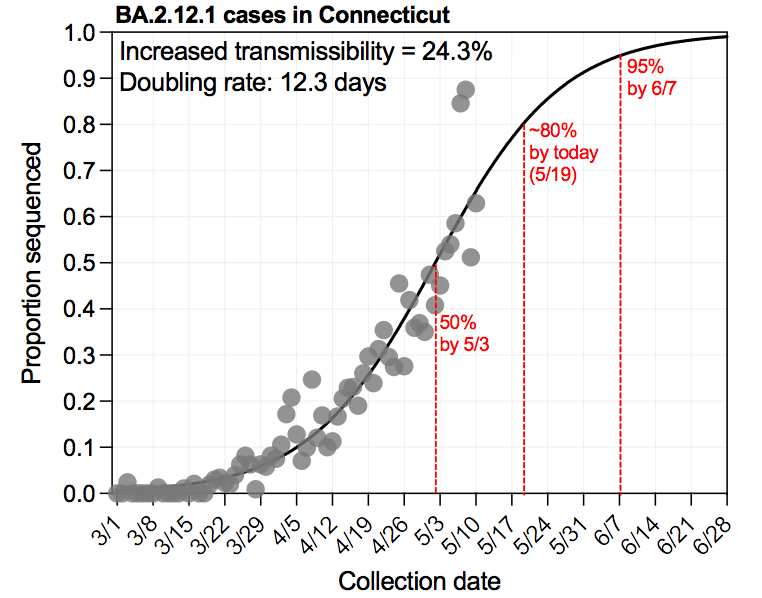





 We are looking into these low level spike amplification samples that should be SGTF to see if this is a lab/TaqPath assay artifact or if there is something about these BA.1 sequences. So far doesn't seem to be sequence-related. Will report (6/16)
We are looking into these low level spike amplification samples that should be SGTF to see if this is a lab/TaqPath assay artifact or if there is something about these BA.1 sequences. So far doesn't seem to be sequence-related. Will report (6/16)



https://twitter.com/NathanGrubaugh/status/1486083856860123137?s=20


 1st, some background
1st, some background








 Here is a link to @maryebushman, @BillHanage et al's paper out in this issue of @CellCellPress (2/16)
Here is a link to @maryebushman, @BillHanage et al's paper out in this issue of @CellCellPress (2/16)

 Our data 👆 do not represent all of Connecticut, but I expect that if Omicron is not dominant yet state-wide, it will be within days.
Our data 👆 do not represent all of Connecticut, but I expect that if Omicron is not dominant yet state-wide, it will be within days. 
 The data are located on our weekly variant report (though I will *try* to update daily). If you scroll down below the summary table, you’ll see the interactive figure. There you can find the data that underline the trends. (2/5)
The data are located on our weekly variant report (though I will *try* to update daily). If you scroll down below the summary table, you’ll see the interactive figure. There you can find the data that underline the trends. (2/5)



 Our data represent a fraction of the samples tested by YNHH, and the catchment is primarily New Haven and Fairfield Counties. So they don’t represent all of Connecticut. I’ve heard from others tracking SGTFs elsewhere in the state that they are not seeing a high % yet. (2/11)
Our data represent a fraction of the samples tested by YNHH, and the catchment is primarily New Haven and Fairfield Counties. So they don’t represent all of Connecticut. I’ve heard from others tracking SGTFs elsewhere in the state that they are not seeing a high % yet. (2/11)



https://twitter.com/NathanGrubaugh/status/1464369814407892996(2/6) The CDC N1 probe mismatch in Omicron is a C28311T mutation. Of the 91 B.1.1.529 sequences on GISAID:


 Likewise, reporting SGTFs will be extremely helpful again for tracking the spread & growth of B.1.1.529, like the amazing folks in South Africa are already demonstrating (2/8)
Likewise, reporting SGTFs will be extremely helpful again for tracking the spread & growth of B.1.1.529, like the amazing folks in South Africa are already demonstrating (2/8)https://twitter.com/Tuliodna/status/1463911577544011780?s=20