⚡️4/8 Connecticut #SARSCoV2 variant surveillance report
@CovidCT | @jacksonlab | @CTDPH | @YaleSPH
Primary news is that B.1.1.7 📈, and we now predict it to be the dominant lineage in CT based on TaqPath SGTF data from Yale + JAX
Short 🧵 | Report 👇
covidtrackerct.com/variant-survei…
@CovidCT | @jacksonlab | @CTDPH | @YaleSPH
Primary news is that B.1.1.7 📈, and we now predict it to be the dominant lineage in CT based on TaqPath SGTF data from Yale + JAX
Short 🧵 | Report 👇
covidtrackerct.com/variant-survei…

2/6 This news follows @CDCDirector Rochelle Walensky saying that B.1.1.7 is "now the most common lineage circulating in the United States".
npr.org/sections/coron…
npr.org/sections/coron…
3/6 We predicted that B.1.1.7 would become dominant in CT in our recent paper led by @tdalpert, @AndersonBrito_, & co. However, we thought that this would have happened earlier in March. The slowdown of B.1.1.7 was likely due to the rapid rise of B.1.526.
cell.com/cell/fulltext/…
cell.com/cell/fulltext/…

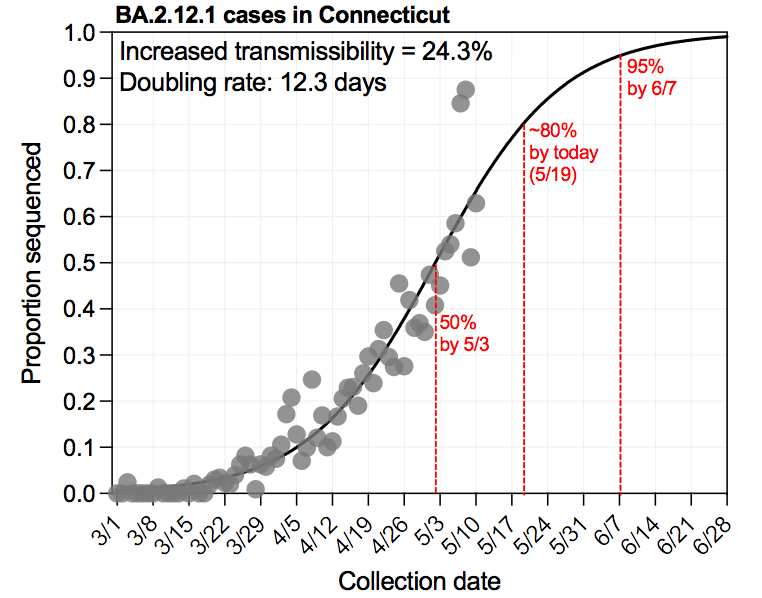
4/6 This figure by @MaryPetrone10 shows how B.1.1.7 and B.1.526 (including B.1.526.1 & B.1.526.2) are increasing at similar rates, displacing most other lineages in CT. Based on our most recent sequencing data, these together make up ~80% of the cases in CT. 

5/6 The co-rise of B.1.1.7 and B.1.526 is leaving little room for other variants - like B.1.351 & P.1 - to emerge and spread. Perhaps this is a good thing, as P.1 + Brazil is a terrible situation, but that really depends on the impacts that B.1.526 will have on public health. 

6/6 This week I'd like to highlight the work by @aewatkins6 who manages a lot of our PCR and lineage reports data. She also has helped to lead many of our NBA/NFL projects prior to this. Here she is with @XtinaHarden and our first delivery of saliva samples from the NBA last June 

• • •
Missing some Tweet in this thread? You can try to
force a refresh