@taekjip is taking over @ApsDbio today to run a tweetorial titled 'single is good but a couple is better'. 

Single molecule methods are allowing direct detection of subpopulations & dynamics, and correlation between multiple observables, with rapidly rising popularity. Technical milestones in single molecule fluorescence can be seen here. 

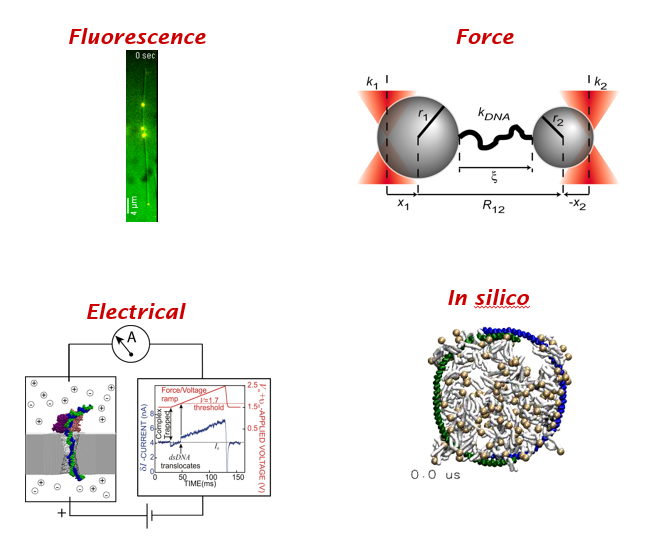
Many flavors of single molecule methods. (1) fluorescence (2) mechanical (3) electrical & (4) in silico. All four have been honored by Nobel prizes in physics, chemistry and physiology. 
For example, the ability to detect single fluorophores is crucial to super-resolution microscopy and single molecule long-read sequencing via zero mode waveguide. See this cool discovery of actin rings in neurons. 

But, going beyond one molecule, one fluorophore or one flavor of single molecule methods can be rewarding. Hence, single is good but a couple is better. As a reminder, you are not fully vaccinated until you get your second shot! 

Single molecule FRET uses dipole-dipole interaction between two fluorophores of different color to infer their relative distance. If you know where in the protein the labels are attached, you can deduce conformational dynamics. See this illustration by Harold Kim.
Moving from a single optical trap to dual optical traps can dramatically improve precision, even allowing us to detect single base pair steps by an RNA polymerase. 

Combining optical trap with single molecule fluorescence can reveal correlation between multiple observables, for example ATP binding and myosin powerstroke as shown here. 

You can even combine ultrahigh resolution dual optical traps with single molecule FRET. See this illustration of ‘fleezers’ by Matt Comstock.
Now a question for you. Can we do something equivalent in single cell studies? Study interaction between two cells in high throughput for example. Regardless, Iet's celebrate single molecule biophysics but better things are ahead when we form a couple. 10/10
• • •
Missing some Tweet in this thread? You can try to
force a refresh