Myself, @GuyBaele & an impressive international cast just got a manuscript up on @medrxivpreprint (medrxiv.org/content/10.110…) about #SARSCoV2 lineage B.1.620 I've tweeted about before. Our findings are a bit alarming & have important implications, so here's a 🧵 1/12
Everything started with a batch of genomes we got back from Lithuanian #SARSCoV2 genomic surveillance on week of Apr 5. We somehow had "B.1.177.57s" with E484K in there. We're not surprised by E484Ks anymore because a small B.1.351 transmission chain crops up occasionally. 2/12
The more we delved the worse it looked:
1. Pangolin was wrong
2. Nextclade indicated lots of private mutations
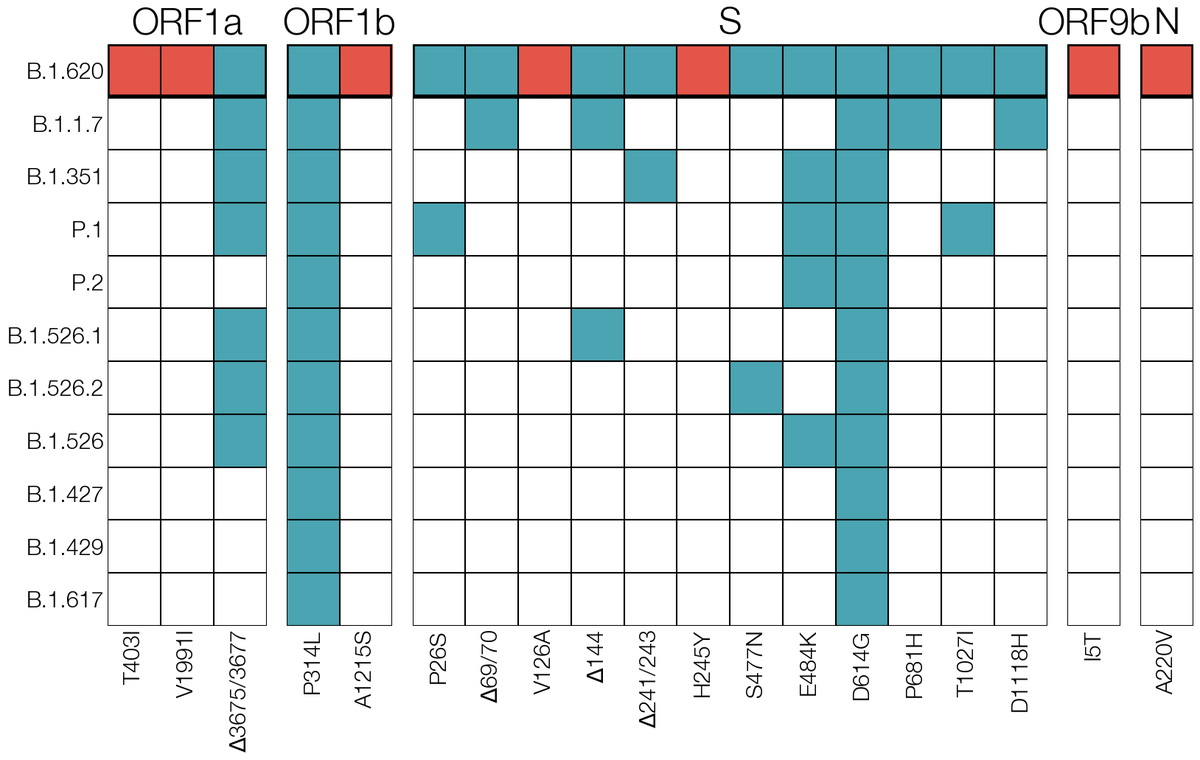
3. Most mutations/deletions in this lineage were seen in VOCs (B.1.1.7, B.1.351, P.1, B.1.526.2)
Always check the mutations, folks. We nearly missed this early. 3/12
1. Pangolin was wrong
2. Nextclade indicated lots of private mutations
3. Most mutations/deletions in this lineage were seen in VOCs (B.1.1.7, B.1.351, P.1, B.1.526.2)
Always check the mutations, folks. We nearly missed this early. 3/12
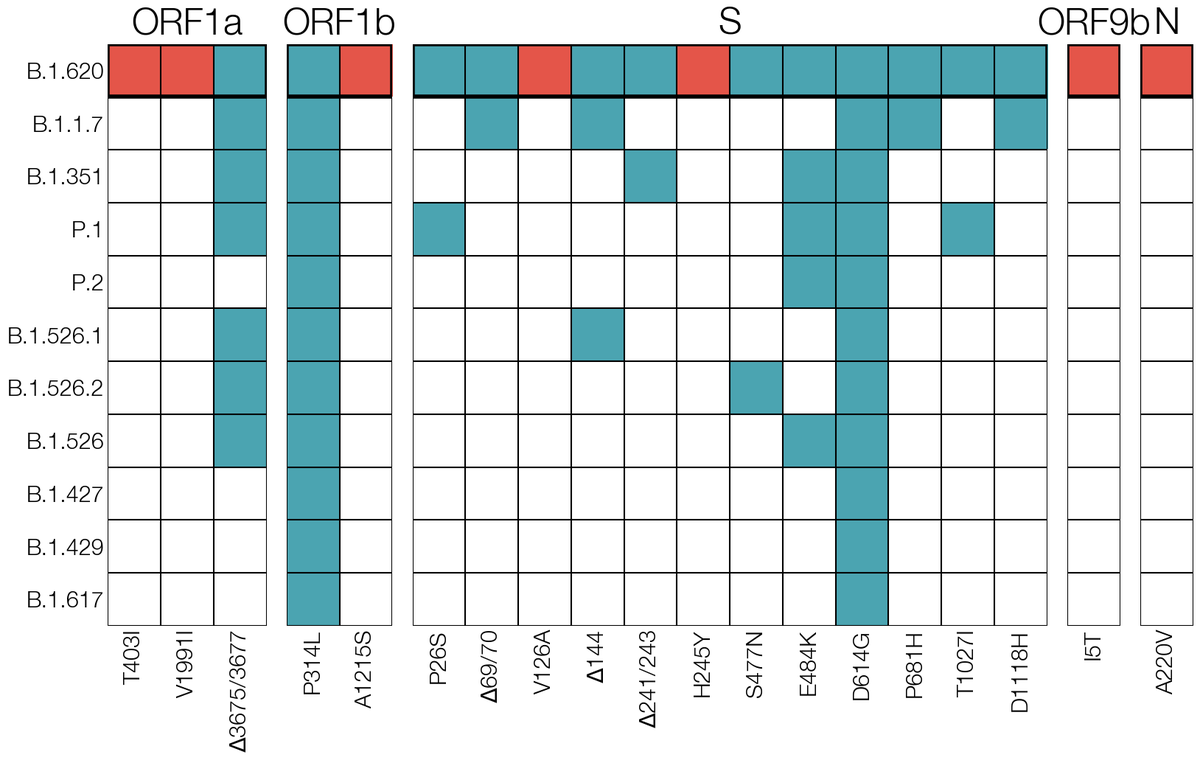
Looking on GISAID for VOC mutations that were never seen in the same background (E484K, S477N & 69del) we found a handful of genomes in France, Belgium, Switzerland, Germany & UK this way. One of them gave us our first clue - one French case returned from Cameroon recently. 4/12
Cue next weeks of frantic emailing anyone who submitted genomes with those mutations to GISAID, a new pango lineage designation request (github.com/cov-lineages/p…) & figure making. We ended up being quite convinced that we were seeing something both concerning & illuminating. 5/12
The connection of this lineage, now designated B.1.620, to Central Africa kept getting stronger as we proceeded - we found more B.1.620 travel cases returning from Cameroon & got unpublished genomes from Central African Republic from collaborators. 6/12 
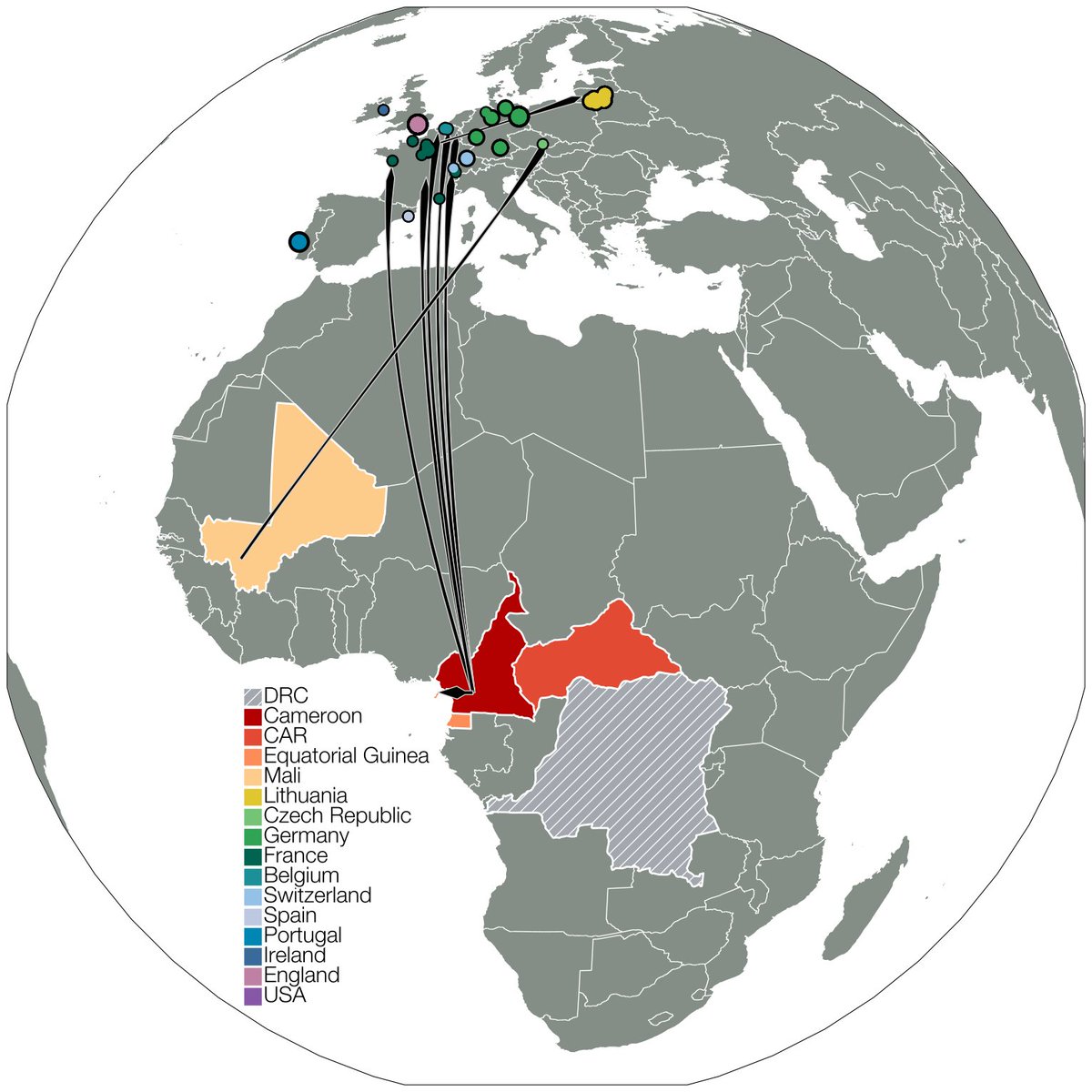
What surprised us is that even then B.1.620 doesn't have close relatives - Swss B.1.619 (also from Cameroon it seems) & some genomes sequenced in Cameroon (thanks to Ahidjo Ayouba) are closest, leaving the B.1.620 branch with ~20 mutations that aren't broken up by relatives. 7/12 
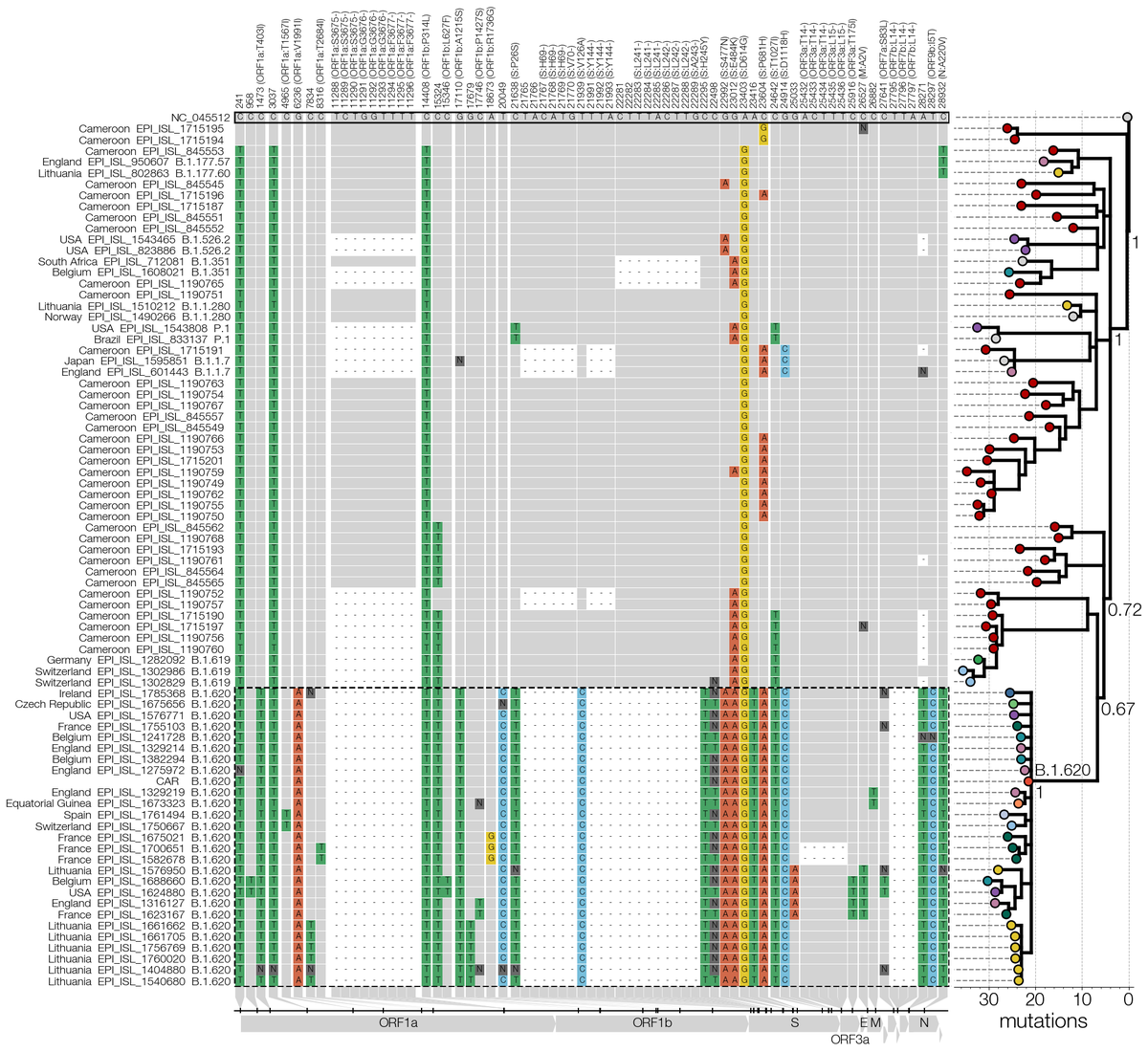
Also worrying is B.1.620's sudden appearance in sequence data - earliest genome we have is Feb 2021. Despite being found on 3 continents all known B.1.620 genomes are genetically homogenous. We think it's common in Central Africa, but it's not doing bad in Europe either. 8/12 
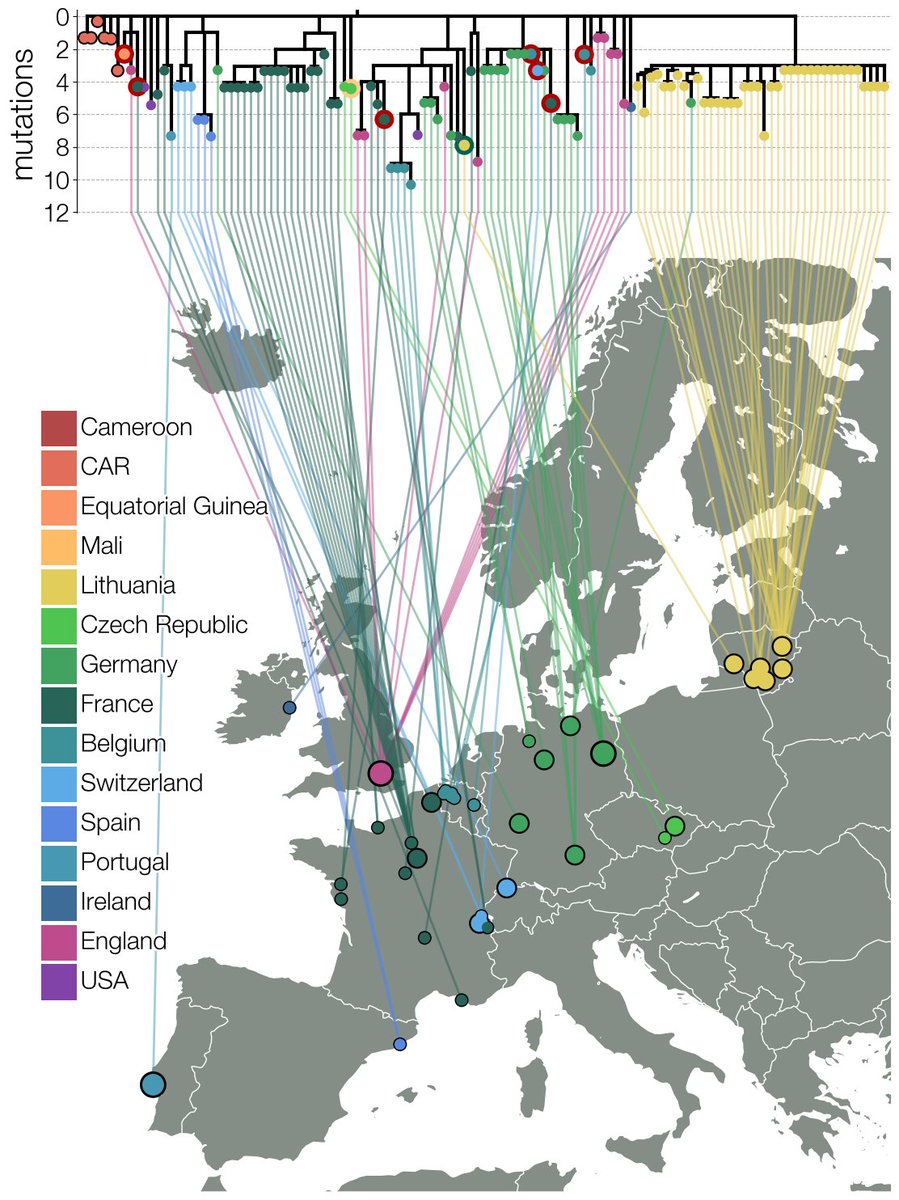
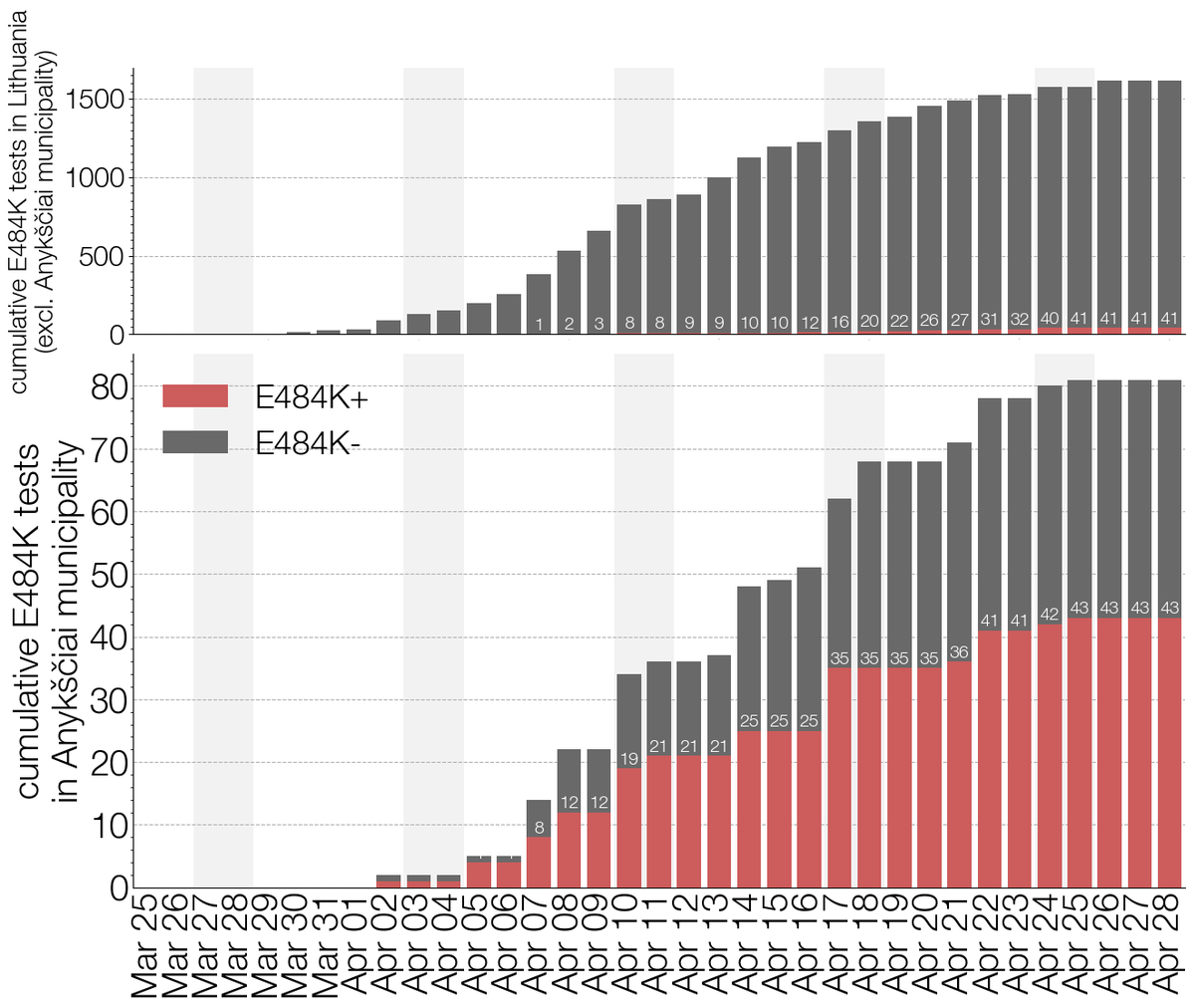
By the time we submitted, B.1.620 continued being found in Europe with new cases cropping up in Ireland, Portugal, Spain & Czechia. In Lithuania B.1.620 continued growing too - a N501Y/E484K/SGTF PCR panel is sometimes used & found >100 cases of B.1.620 across the country. 9/12 
To sum up B.1.620 is a #SARSCoV2 lineage that appeared suddenly with numerous concerning mutations, no known close relatives & is probably common in parts of Africa & is now found in Europe. But consider the implication of this - what if B.1.620 is the tip of the iceberg? 10/12
International organisations don't provide enough sequencing & vaccines to countries that can't get their own & we don't know what #SARSCoV2 variants are selected after more than a year of unimpeded circulation. If you don't sequence you will never know & everyone loses. 11/12
Limited ongoing sequencing already gave us the "Tanzania variant", A.23.1 & now B.1.620. What else is out there? How bad is it likely to be? We can't tell. Infectious diseases are rarely just one country's issue. #SARSCoV2 is a global problem, its solution has to be too. 12/12
• • •
Missing some Tweet in this thread? You can try to
force a refresh