
We often discuss how more comprehensive and sensitive techniques improve the diagnostic yield for patients affected by rare genetic diseases. Indeed, yields have improved as we've gone from microarrays to whole genome #sequencing.
However, there's another critical component.
However, there's another critical component.
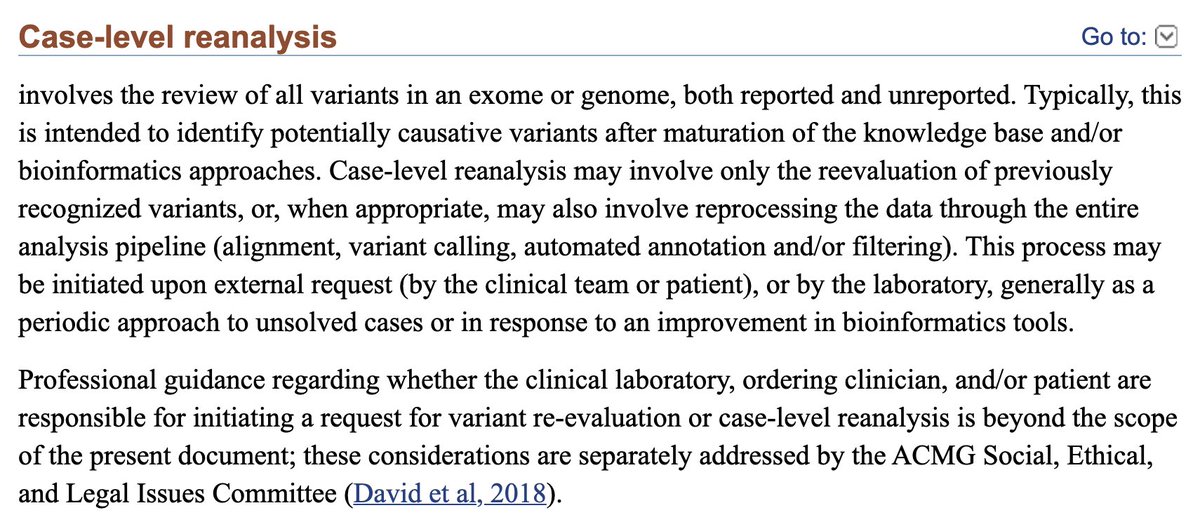
Case-Level Reanalysis (CLR)
By reanalyzing genomic data, as our global knowledge-base grows, we improve diagnostic yields.
We believe the broadest tests should be done first to avoid the need to re-contact and re-accession patient samples.
ncbi.nlm.nih.gov/pmc/articles/P…
By reanalyzing genomic data, as our global knowledge-base grows, we improve diagnostic yields.
We believe the broadest tests should be done first to avoid the need to re-contact and re-accession patient samples.
ncbi.nlm.nih.gov/pmc/articles/P…
The economics for both the lab (and patient) change dramatically as well in a 'generate-once-reassess-often' framework. As more is known, variant interpretation may shift from being more manual to more automated.
Still, this is a really hard technological problem.
Still, this is a really hard technological problem.
Variant interpretation, we think, is the most difficult and least commoditized aspect of sequence data analysis. It also follows the garbage-in-garbage-out rule. This is another reason why we believe rare disease patients should receive comprehensive sequencing.
As the technology improves on the bases of cost and feasibility, we think phased genome assembly is the most powerful tool for comprehensive variant detection, especially for patients afflicted with rare disease.
https://twitter.com/GenomeInABottle/status/1402056739701379082
• • •
Missing some Tweet in this thread? You can try to
force a refresh




