We have just released a new version of the Nextclade web application clades.nextstrain.org for #SARSCOV2. We now use Nextalign (C++ via wasm) under the hood, which allows for many new exacting features!
A quick tour!
1/7
A quick tour!
1/7

Analysis now takes a few seconds to start, but overall it has become much faster and analyzing hundreds of sequences should be no problem. You can download the reference alignments, including translations of SARS-CoV-2 genes, for further analyses.
2/7
2/7

You can now examine the diversity of the nucleotide alignment and alignments of different proteins. Select a gene from the drop-down or click on a gene in the genome annotation panel below.
3/7
3/7
The tooltips to explore diversity have become much more informative. For amino acid changes, we now provide a nucleotide context view that is particularly helpful for complex mutations. Consecutive changes are merged into one tooltip.
4/7
4/7

A little icon next to the sequence tells you whether some genes of the sequence have failed to translate, either because they weren't covered or because of frame shifts.
5/7
5/7

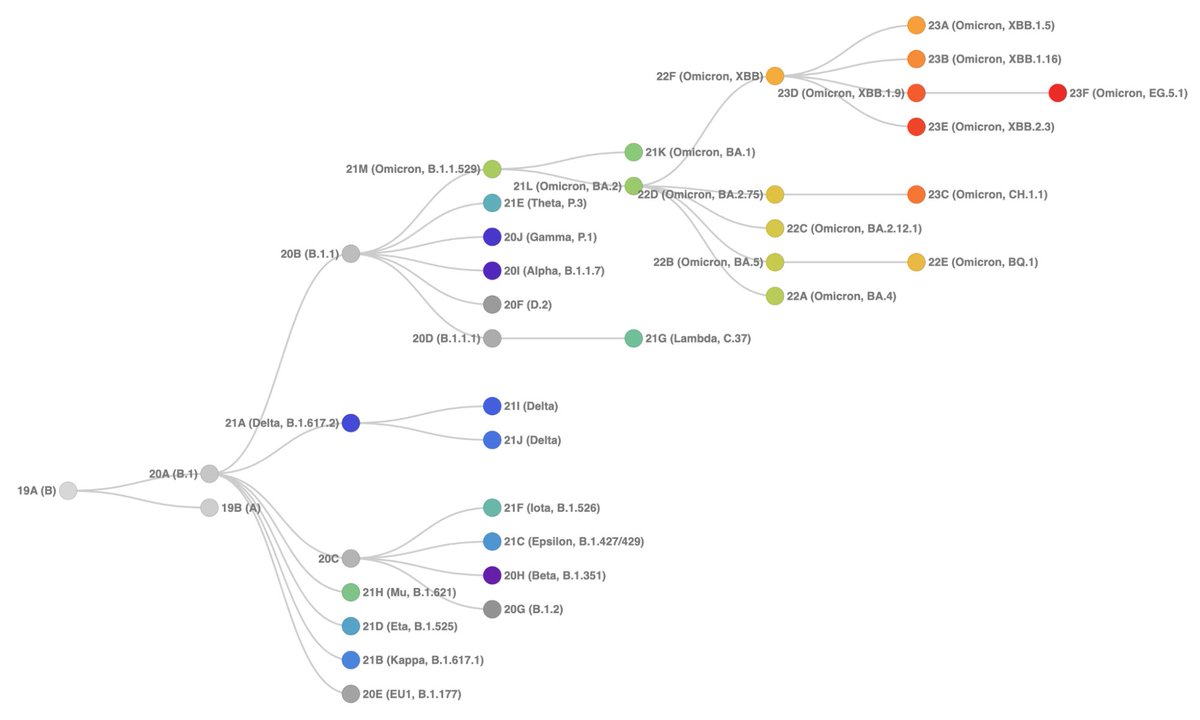
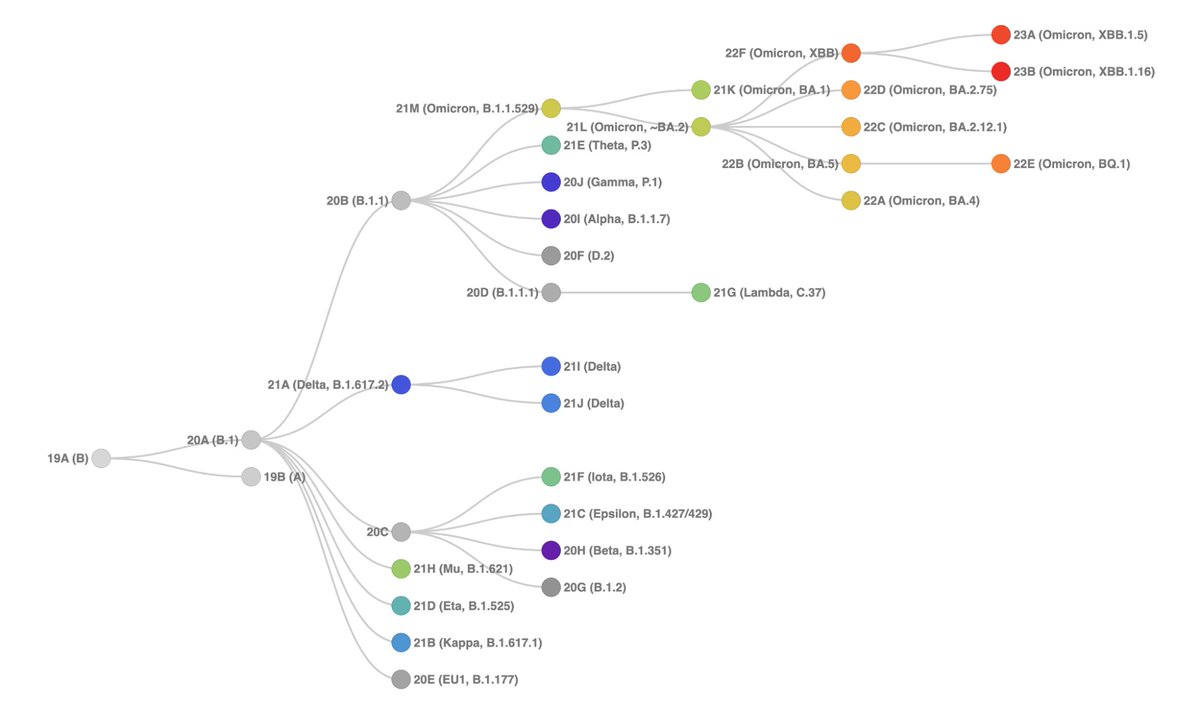
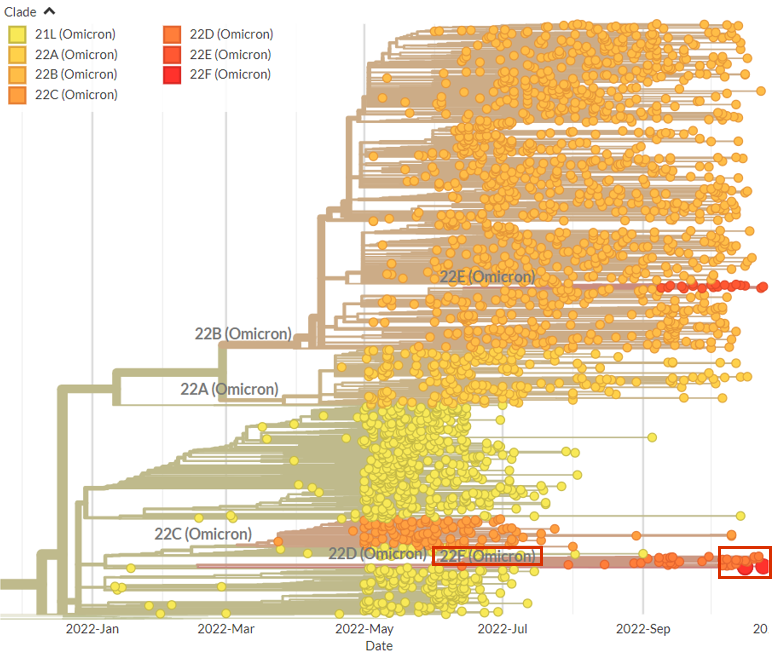
As before, the samples are placed on a reference tree that now includes the updated #Nextstrain clades and a diverse set of lineages.
6/7
6/7

Brought to you by the @nextstrain team.
Share your comments and suggestions at discussion.nextstrain.org or github.com/nextstrain/nex… or reach out to @ivan_aksamentov.
7/7
Share your comments and suggestions at discussion.nextstrain.org or github.com/nextstrain/nex… or reach out to @ivan_aksamentov.
7/7
• • •
Missing some Tweet in this thread? You can try to
force a refresh