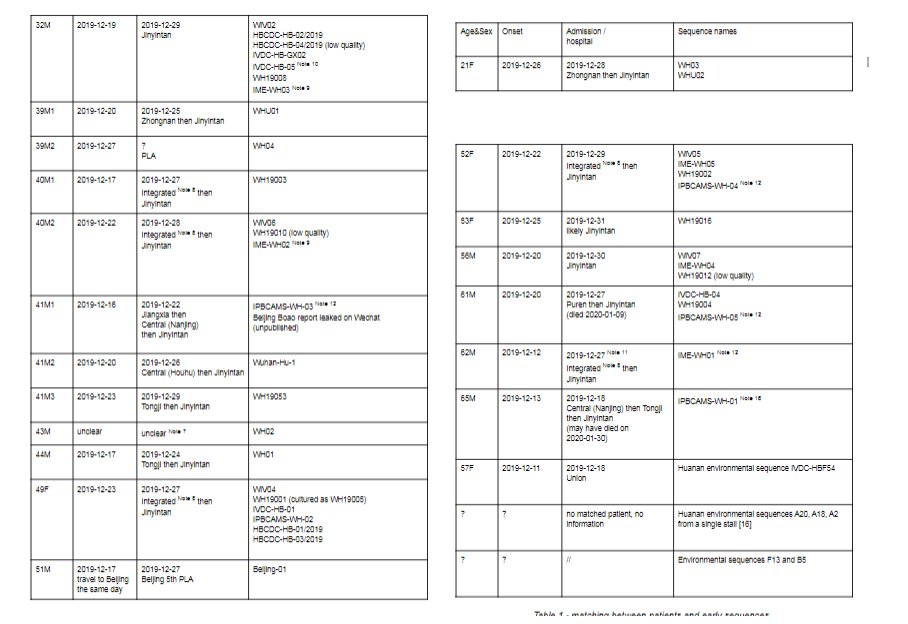
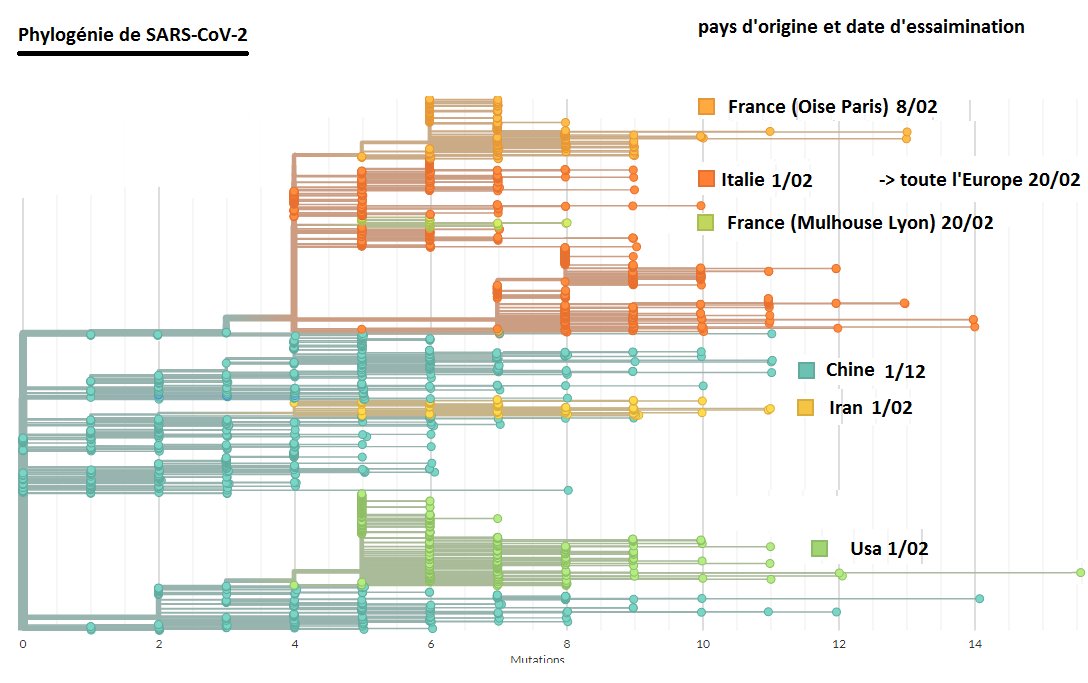
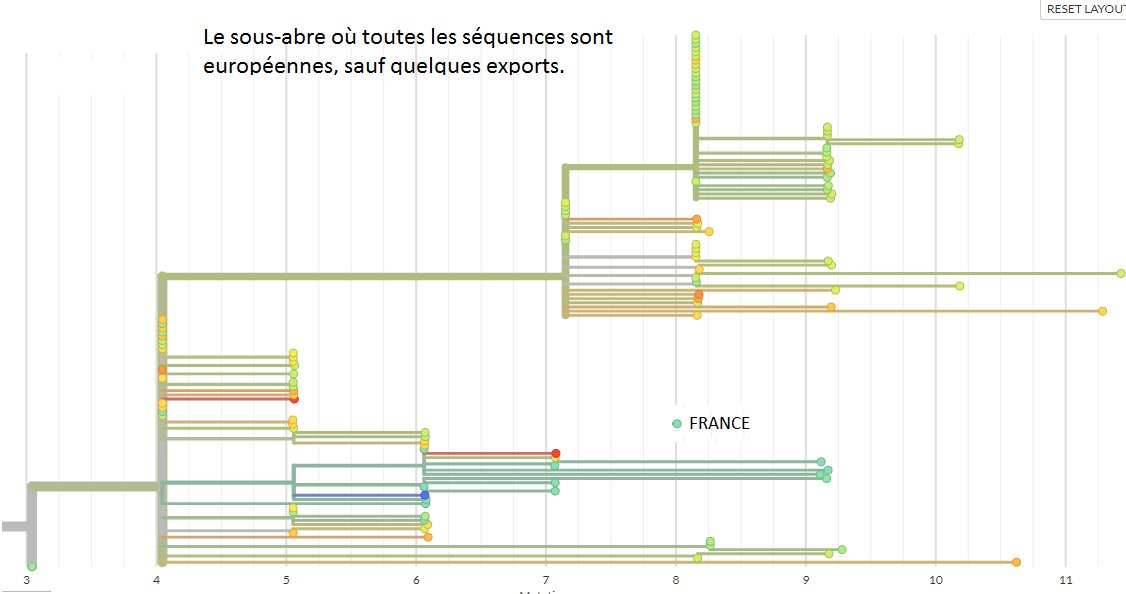
An update on my partial RdRp #sarbecovirus tree. This is now the region 15000-16000 with many smaller sequences.
nextstrain.org/community/baba…
Once sample is collected they do a PCR, if positive they sequence the region 15300-15700, if interesting they try to sequence the whole genome

nextstrain.org/community/baba…
Once sample is collected they do a PCR, if positive they sequence the region 15300-15700, if interesting they try to sequence the whole genome


Species: Rhinolophus, 9 Hipposideridae, 2 Human, 2 Civet, 3 Pangolin, 1 Chinese badger, 2 Vesper, 1 Molossidae.
No rhinolophids in America. In other countries rhinolophids were sampled and were negative to CoV or sarbecovirus. Turkey unsampled, there are sarbecoviruses there!
No rhinolophids in America. In other countries rhinolophids were sampled and were negative to CoV or sarbecovirus. Turkey unsampled, there are sarbecoviruses there!

Recently published:
Lebanon, Russia (Sotchi), Croatia, UK.
In the SARS 1&2 clades: the Menglun samples, Thailand,Cambodia, 12 sequences likely from Jinning (where almost all good matches to SARS-CoV come from).
The African lineage was seen only in the great lakes region.
Lebanon, Russia (Sotchi), Croatia, UK.
In the SARS 1&2 clades: the Menglun samples, Thailand,Cambodia, 12 sequences likely from Jinning (where almost all good matches to SARS-CoV come from).
The African lineage was seen only in the great lakes region.

Four clades:
🔹Europe-Africa
🔹SARS2 (Yunnan)
🔹SARS (Yunnan)
🔹HKU3-Rf1 (Hong-Kong,Hubei,Zhejiang,Jilin,Guizhou..)
🔹Unclassified: India, Japan, a small Yunnan lineage.
🔹Outlier: Taiwan, in the Europe-Africa clade (?)


🔹Europe-Africa
🔹SARS2 (Yunnan)
🔹SARS (Yunnan)
🔹HKU3-Rf1 (Hong-Kong,Hubei,Zhejiang,Jilin,Guizhou..)
🔹Unclassified: India, Japan, a small Yunnan lineage.
🔹Outlier: Taiwan, in the Europe-Africa clade (?)

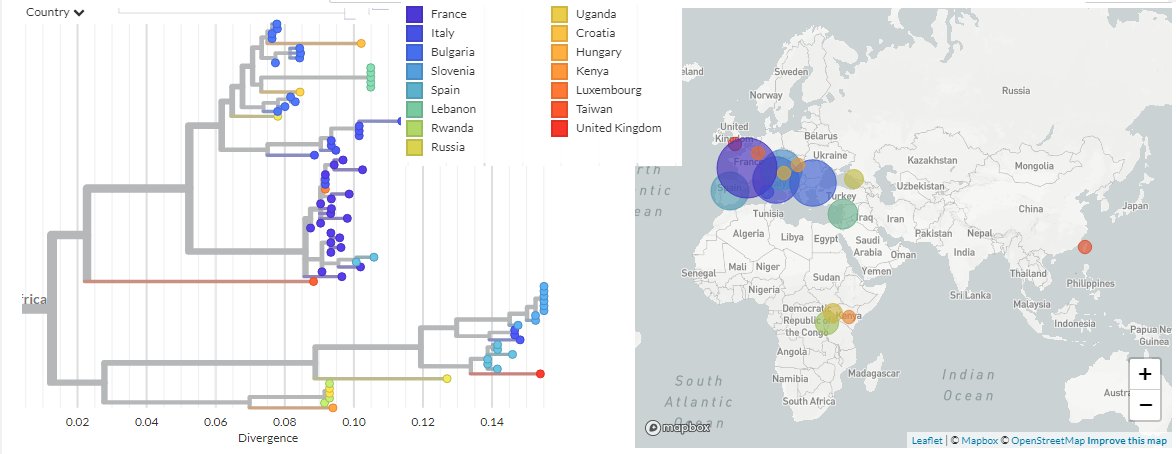
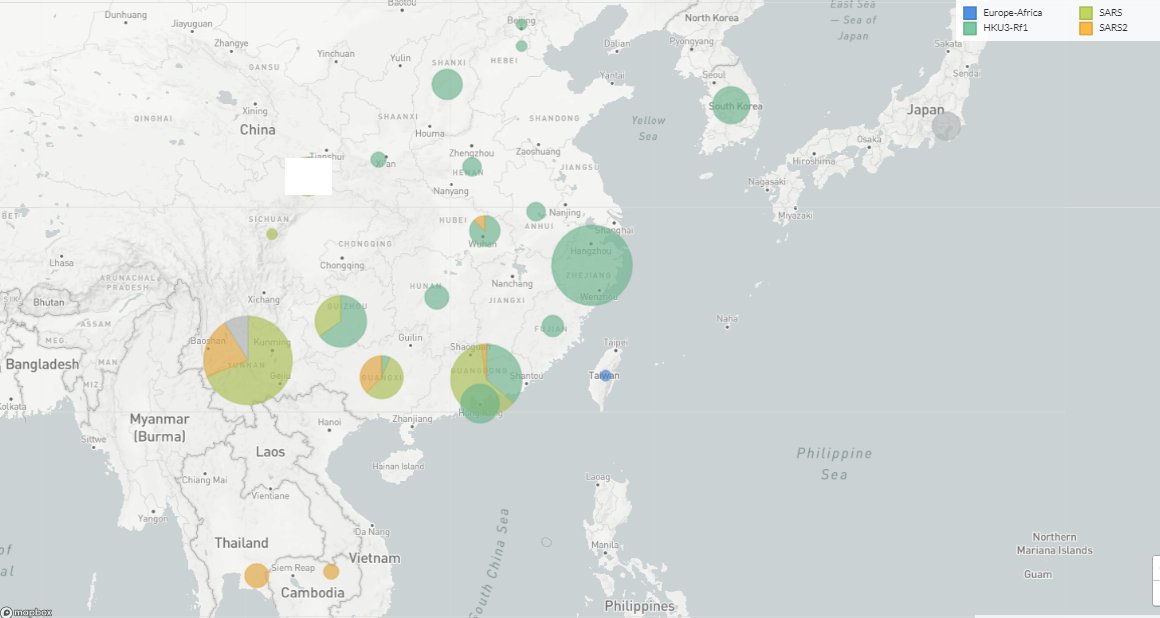
About the spillover potential: overall it is likely low, see all the species sampled in relation to the PREDICT project in a rectangle containing Yunnan and its borders, most (non-Rhinolophus) samples were tested for CoV and didn't contain sarbecoviruses. 



• • •
Missing some Tweet in this thread? You can try to
force a refresh