
Excited to share our new #method, scDRS, for associating **individual cells** to disease by integrating #scRNA-seq + #GWAS, providing #polygenic disease risk enrichment beyond tissues or cell types.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
scDRS assesses excess expression of a set of putative disease genes constructed from GWAS, using an appropriately matched empirical null distribution to compute p-values. (2/n) 

We applied scDRS to GWAS from 74 diseases and complex traits (average N=341K) in conjunction with 16 scRNA-seq data sets spanning 1.3 million cells from 31 tissues and organs, including the amazing Tabula Muris Senis mouse atlas and Tabula Sapiens human atlas @czbiohub. (3/n) 

Results at cell type-level recapitulated known biology. Importantly, many associated cell type-disease pairs showed significant heterogeneity (247 of 577 pairs). (4/n) 

Individual cell-level results include subsets of T cells assoc. with IBD characterized by their effector-like states; pyramidal neurons assoc. with SCZ characterized by their spatial location; and hepatocytes assoc. with TG characterized by their higher ploidy levels. (5/n) 
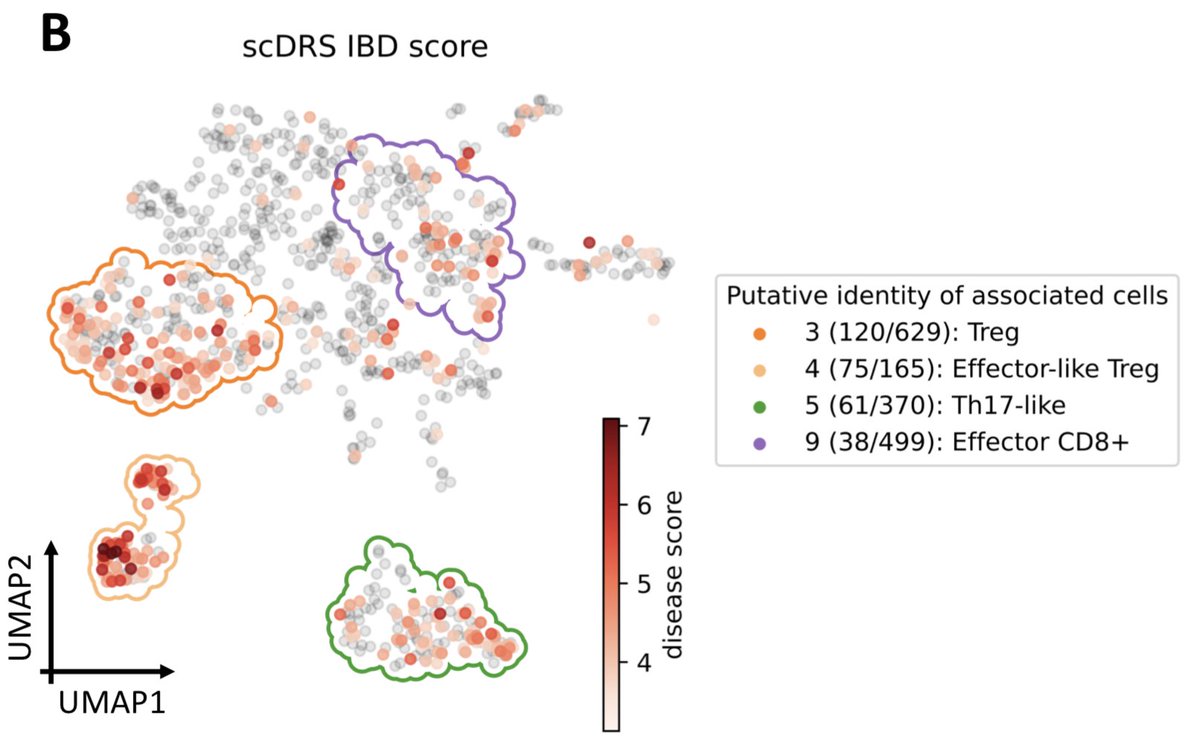
Genes whose expression across individual cells was correlated with the scDRS score (thus reflecting co-expression with GWAS disease genes) were strongly enriched for gold-standard drug target and Mendelian disease genes. (6/n) 

Co-led by @kangchenghou and supervised by @bpasaniuc and Alkes Price. Many thanks to our amazing collaborators: @soumya_boston, @mikejg84, Bruce Wang, @james_y_zou, @drAOPisco, @kanishkadey, @_kjag, Katy Weinand, @saorisakaue, Aris Taychameekiatchai, and Poorvi Rao (7/7)
• • •
Missing some Tweet in this thread? You can try to
force a refresh



