And now, a mega-thread: If you've ever wondered what connects all our work at @viralemergence, our new paper in @NatureMicrobiol ties it all together. No, really. It's all one thing. Want to step through the Verena Cinematic Universe together? 

Our team uses big data, statistics, and machine learning to understand "the science of the host-virus network", a broad methodological problem that includes a number of smaller, more applied problems.
nature.com/articles/s4156…
nature.com/articles/s4156…

If you want to study the host-virus network, you need data. But as @roryjgibb & co. showed, existing datasets are full of taxonomic inconsistencies and conflicts. We needed a synthesis.
academic.oup.com/bioscience/art…

academic.oup.com/bioscience/art…


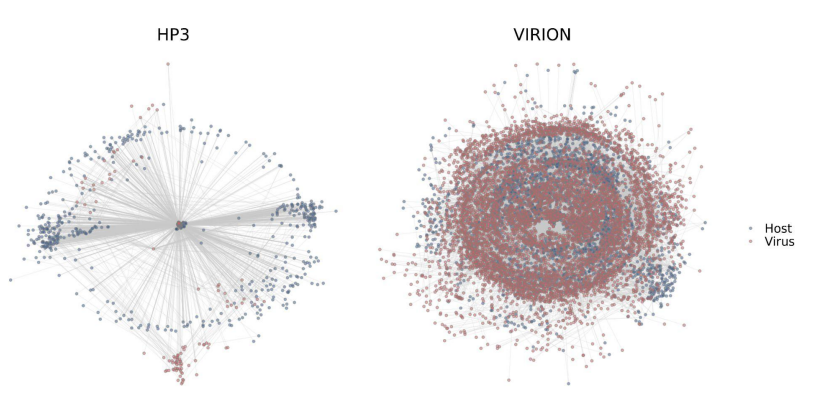
That's why we built Global Virome in One Network (VIRION), the most comprehensive (and clean) atlas of the vertebrate virome ever developed.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
Our work doesn't stop there; @taddallas & co. built a new interface to the Ecological Database of the World's Insect Pathogens, one of the few resources available for the insect-pathogen network.
ecoevorxiv.org/yd3x5/
ecoevorxiv.org/yd3x5/
What can we use those kinds of data to do?
(1) Predict the host-virus network!
@tpoi & co. propose a new method to recover the "missing" network (>90% of it), especially in the Amazon basin, which has been severely undersampled for wildlife viruses.
arxiv.org/abs/2105.14973

(1) Predict the host-virus network!
@tpoi & co. propose a new method to recover the "missing" network (>90% of it), especially in the Amazon basin, which has been severely undersampled for wildlife viruses.
arxiv.org/abs/2105.14973


What makes models like this work? @taddallas and @danjbecker showed that both phylogenetic and geographic scale can drive variation in model performance:
cambridge.org/core/journals/…
cambridge.org/core/journals/…

@taddallas & co. have also recently shown that you can extend the host-virus network problem to predict the host-vector-virus network for mosquito-borne flaviviruses
ecoevorxiv.org/xzmp8/


ecoevorxiv.org/xzmp8/



You can read more about the theory of predicting ecological networks over space and time in this amazing team review organized by @tpoi & co.:
royalsocietypublishing.org/doi/full/10.10…

royalsocietypublishing.org/doi/full/10.10…

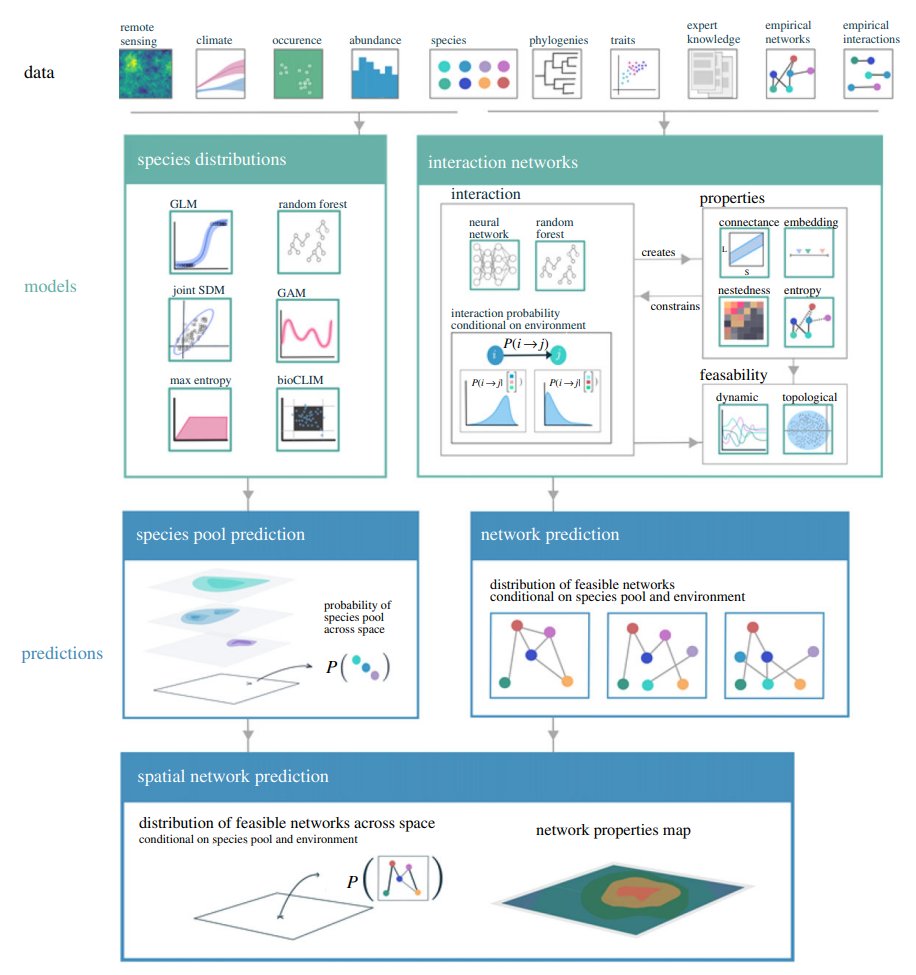
(2) Predict which viruses can infect humans.
We've been able to use our host-virus network predictions + network embedding to create the most accurate artificial intelligence ever developed for zoonotic risk prediction. No, really.
arxiv.org/abs/2105.14973

We've been able to use our host-virus network predictions + network embedding to create the most accurate artificial intelligence ever developed for zoonotic risk prediction. No, really.
arxiv.org/abs/2105.14973


The backbone of that model was just published by @NardusMollentze with his own group, and I can't recommend this paper strongly enough. A masterpiece of predictive research.
journals.plos.org/plosbiology/ar…
journals.plos.org/plosbiology/ar…

We've also written as a team about where tech like this might start to be useful for global health security, building on a workshop that brought together bench virologists, computational experts, and global health practitioners.
royalsocietypublishing.org/doi/full/10.10…
royalsocietypublishing.org/doi/full/10.10…

In the future, it'll be important to go beyond zoonotic potential and also look at epidemic potential, including transmissibility...
journals.plos.org/plosone/articl…
journals.plos.org/plosone/articl…
...and virulence.
pnas.org/content/116/16…
pnas.org/content/116/16…
(3) Predicting viral reservoirs.
I should say - we owe so much on this to @bahanbug, who developed the foundational approach: identify what animals have some viruses of interest; use machine learning to predict which others might also have those viruses.
journals.plos.org/plosntds/artic…
I should say - we owe so much on this to @bahanbug, who developed the foundational approach: identify what animals have some viruses of interest; use machine learning to predict which others might also have those viruses.
journals.plos.org/plosntds/artic…
At the start of the pandemic, our team (led by @danjbecker and @Gfalbery) built the first multi-model comparison study to predict potential bat reservoirs of undiscovered betacoronaviruses.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…

We've spent the last two years proving that we can, actually, use artificial intelligence to optimize the search for new betacoronaviruses. Now we know that they work best when we actually use data on species "traits" - their ecology, evolution, immunology. 

We also increasingly think that these predictions can be improved by using data that better represent reservoir competence for viruses (e.g., viral isolation instead of just PCR or serology)
cell.com/trends/ecology…
cell.com/trends/ecology…

In some of our collaborative work with the Forbes lab predicting rodent reservoirs of undiscovered orthohantaviruses in the Americas, we see that improvement when we model viral isolation vs. PCR:
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
We also think tools like this will increasingly help us anticipate pathogen "spillback" from humans into wildlife. This review by @annafagre & co. outlines the theory behind the approach, as well as the data we'll need...
ecoevorxiv.org/sx6p8/
ecoevorxiv.org/sx6p8/

...and this new study by @bahanbug's group, which uses ACE2 sequences to predict potential wildlife hosts of SARS-CoV-2, is proof-of-concept that this could work!
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
4. Viral diversity: why do some animals have more viruses? Why do they have more *zoonotic* viruses?
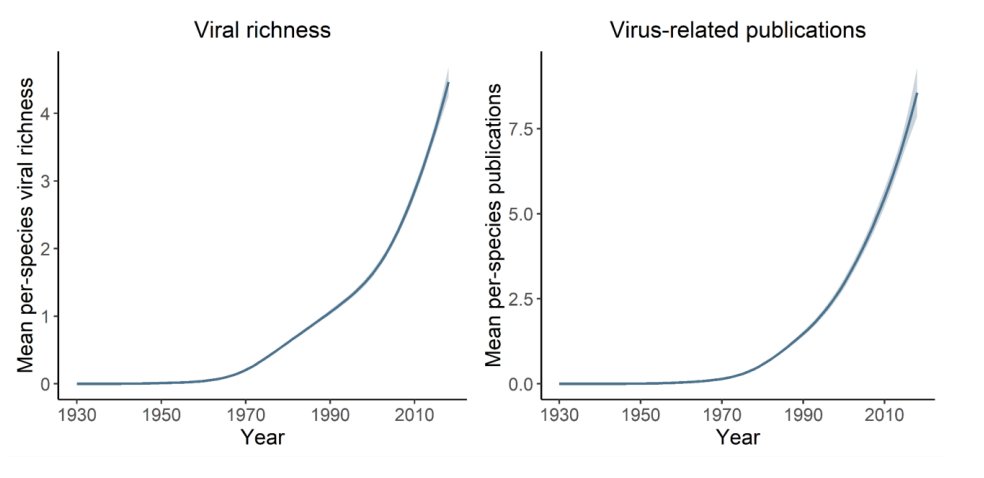
Things get a bit complicated here. In some of my own work we've shown that we probably only really know about ~1% of the mammal virome (50k+ viruses).
nature.com/articles/s4155…
Things get a bit complicated here. In some of my own work we've shown that we probably only really know about ~1% of the mammal virome (50k+ viruses).
nature.com/articles/s4155…
@roryjgibb & co. showed that this creates a bit of a problem: where we look for viruses, we find them. And if we ask questions about viral diversity, the answers change over time.
biorxiv.org/content/10.110…

biorxiv.org/content/10.110…

@Gfalbery & co. showed that some key ecological hypotheses don't hold up after you correct for sampling bias. Sure, animals in cities have more known zoonotic viruses - because they have more known viruses, because we're looking more.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…

Results like these are an unexpected challenge to the "pace-of-life theory" - i.e., the idea that fast-lived species like rodents make immune investments and invade habitats in ways that predispose them to zoonotic risk. @Gfalbery and @danjbecker explain:
cell.com/trends/parasit…
cell.com/trends/parasit…
@Gfalbery @danjbecker We've also got some preliminary results that take this further, showing that domestication probably increases zoonotic viral richness, but involvement in the wildlife trade doesn't.
github.com/viralemergence…
github.com/viralemergence…

@Gfalbery @danjbecker 5. Which hosts share viruses with each other?
@Gfalbery & co. showed that - like dozens of studies have shown at smaller scales - it's your proximity to other animals in geographic space and evolutionary time that determines how similar your viruses are
nature.com/articles/s4146…
@Gfalbery & co. showed that - like dozens of studies have shown at smaller scales - it's your proximity to other animals in geographic space and evolutionary time that determines how similar your viruses are
nature.com/articles/s4146…

@Gfalbery @danjbecker This might seem like well-covered ground, but there's tons of questions left to answer. Heck, a new paper in @NatureMicrobiol shows that the same principles govern gut archaea across vertebrates:
nature.com/articles/s4156…
nature.com/articles/s4156…
@Gfalbery @danjbecker @NatureMicrobiol Together, @Gfalbery and I have applied the same viral sharing model to predict how climate change-driven range shifts could completely reshape the global virome, creating hotspots of cross-species transmission in the places where we'll live in 2050
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…

@Gfalbery @danjbecker @NatureMicrobiol I'm really not kidding when I say "it's all one thing": predicting the next pandemic virus, tracing SARS-CoV-2 to its origin, projecting hotspots of climate-driven disease risk - it's all the science of the host-virus network. That's what we do.
nature.com/articles/s4156…
nature.com/articles/s4156…
• • •
Missing some Tweet in this thread? You can try to
force a refresh