
There have been a number of overview threads on the emerging variant designated as @PangoNetwork lineage B.1.1.529, @nextstrain clade 21K and @WHO Variant of Concern Omicron. I'm not going to attempt to be comprehensive here, but will highlight a few aspects of the data. 1/16
Global systems for identifying novel variants and rapidly sharing data are working well with 91 genomes from Omicron viruses shared to @GISAID from specimens collected between Nov 11 and Nov 23 from Botswana, South Africa and Hong Kong. 2/16
These viruses are visible on @nextstrain as "21K (Omicron)" shown here in red (nextstrain.org/ncov/gisaid/af…). They do not descend from previously identified "variant" viruses and instead their closest evolutionary connection is to mid-2020 viruses. 3/16 
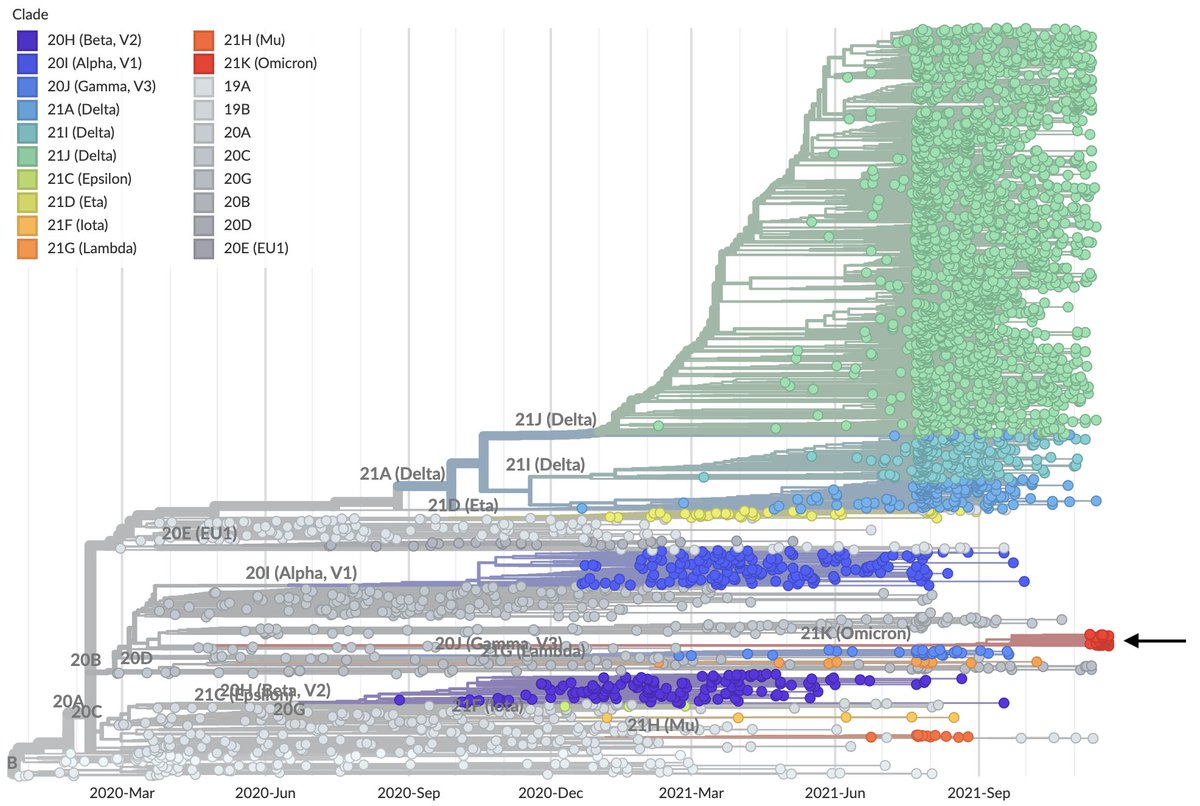
This extremely long branch (>1 year) indicates an extended period of circulation in a geography with poor genomic surveillance (certainly not South Africa) or continual evolution in a chronically infected individual before spilling back into the population. 4/16 

This long branch does not appear to be an artifact of subsampling. A CoVariants "focal build" that pulls in related genomes also shows this long branch (nextstrain.org/groups/neherla…) as does a search with UShER (genome.ucsc.edu/cgi-bin/hgPhyl…) for placement on the full SARS-CoV-2 tree. 5/16 

As noted by @Tuliodna these viruses bear a remarkable constellation of mutations in the spike protein that are concerning in terms of predicted immune escape coupled with increased transmissibility. 6/16
https://twitter.com/nicd_sa/status/1463833794276900864
You can see the striking accumulation of mutations in the S1 domain of the spike protein just by plotting S1 mutations against time and coloring by clade (nextstrain.org/ncov/gisaid/af…). Omicron viruses bear many more S1 mutations than previously circulating variants. 7/16 

Besides this mutational profile, the other critical data point is apparent rapid spread in South Africa. Here, I wouldn't trust frequencies of genome data at the moment as there has been urgent sequencing and deposition of S dropout samples collected from Gauteng. 8/16
This causes a straight forward genome frequencies analysis to overestimate rate of spread of Omicron. Instead I'd focus on rapid increase in proportion of S dropout tests in Gauteng as well as other provinces in South Africa (screen grab from ). 9/16 

This rapid increase across provinces is concerning and suggests that Omicron is outcompeting Delta in the current context in South Africa. I'd hope for modeling work using this S dropout data to better quantify relative fitness of Omicron vs Delta in the following days. 10/16
However, rather than looking at raw frequency of genomic lineage or clade assignments, we can take a phylodynamic approach to estimate clade growth. This approach uses a molecular clock along with branching patterns in the phylogeny to infer viral population dynamics. 11/16
We did this previously for early data in Washington State to infer the introduction time and rate of exponential growth when community transmission was first identified in Feb/Mar 2020 (Figure 2B of bedford.io/papers/bedford…). 12/16 

Here, I took the exact same approach using 77 available Omicron genomes from South Africa and Botswana. This yields a median estimate of a common ancestor at Oct 7 (95% CI between Sep 19 and Oct 21). This seems consistent with first detection in a sample from 11 Nov. 13/16 

This also yields a median estimate of exponential doubling time of 4.8 days (95% CI between 2.6 and 8.7 days). This median estimate can be compared to a 3.4 doubling time for early spread in Washington State. 14/16 

Growth rate (in absolute terms and relative to Delta) will be become clearer in the following days, but at the moment, I believe we're looking at a variant that potentially has significant immune evasion and that appears to be spreading rapidly. 15/16
The world should be immensely grateful to @Tuliodna, @ceri_news, @nicd_sa, the Network for Genomics Surveillance in South Africa and the Botswana Harvard HIV Reference Laboratory for discovering this variant and immediately alerting to its existence. 16/16
• • •
Missing some Tweet in this thread? You can try to
force a refresh







