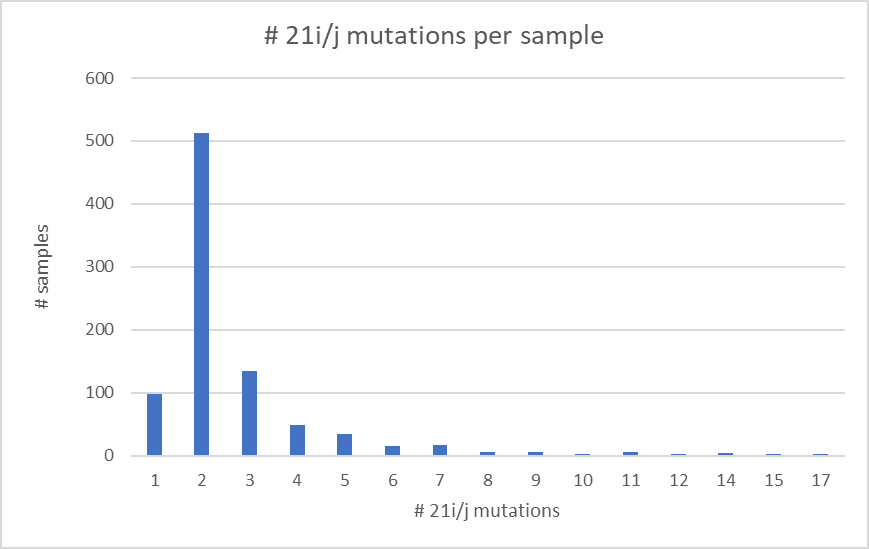
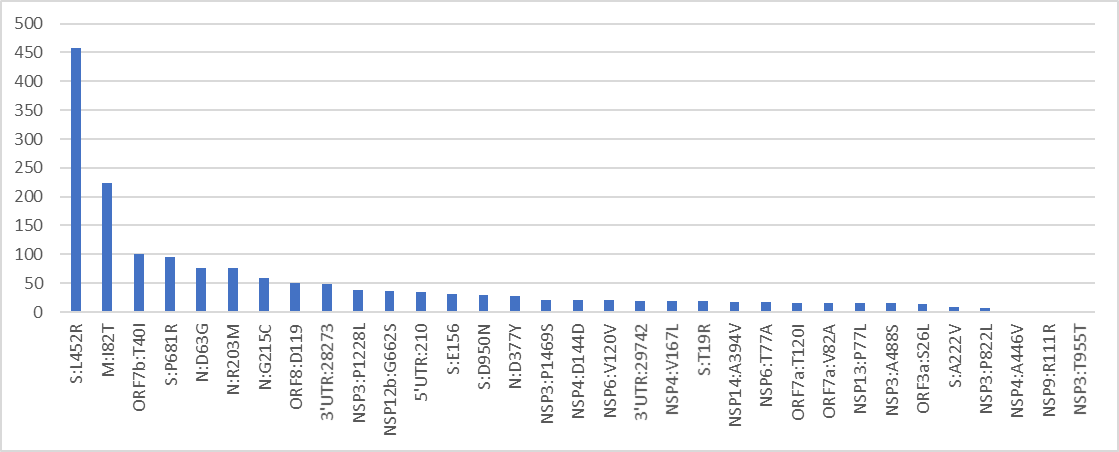
Omicron is a weird variant.
Together with @SternLab i took a dive into it's strangeness from the genomic point of view by analyzing the defining mutations of the 3 Omi’s (BA.1/2/3) and 65 other variants having a long branch in the tree and high % of NS mutations.
Together with @SternLab i took a dive into it's strangeness from the genomic point of view by analyzing the defining mutations of the 3 Omi’s (BA.1/2/3) and 65 other variants having a long branch in the tree and high % of NS mutations.
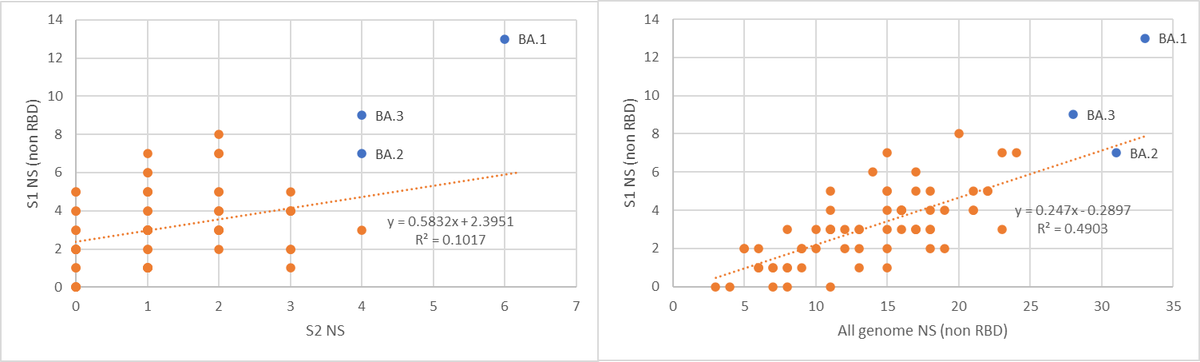
The strangest thing of course is the RBD mutations. There are a lot. As seen, they are not in line with the trend of many other variants. 
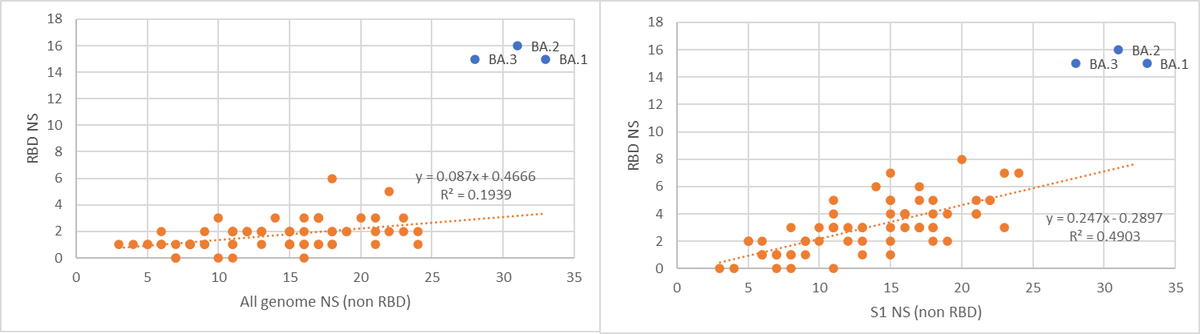
So, as we know from the last year, mutations in RBD evolved quite clear “boundaries”. In the other 3 VOC’s there were no more than 3 non-synonymous RBD mutations. In Omi, it seems that there are new “boundaries” from some reason (more on that in the end of this thread).
Beyond S-
In the 65 variants we compared to, 24 had one mutation in M gene and only 1 had two (B.1.632). Omi has three, two of which are completely unique.
In the 65 variants we compared to, 24 had one mutation in M gene and only 1 had two (B.1.632). Omi has three, two of which are completely unique.
Deletions in NSP6 are also something quite convergent. 21 variants has the deletion in positions 106-108 (SGF), including BA.2&3 . 2 Variants has single AA deletion in position 104. And only one variant (B.1.1.524) shares the 105-107 deletion of Omi.
So, something in the mutation profile, especially in the RBD, seems to be not in line with the general trend. It’s not just another variant but one with a lot more mutations, it’s different.
The 2 main scenarios for Omi’s evolvement are chronic patient/’s or reverse zoonosis followed by another spillover to humans.
The reverse zoonosis may be less likely as there 3 different variants in the Omi lineage, one is a recombinant of the other 2. All arose in the same time.
So a chronic patient/’s with several variants evolved in (co-circulating,making recombination probable) seems more likely.
So a chronic patient/’s with several variants evolved in (co-circulating,making recombination probable) seems more likely.
But, as @sergei pond demonstrated, many mutations fixed in Omi were previously shown to be under negative selection. Epistasis is a possible explanation.
https://twitter.com/sergeilkp/status/1468702270942584832
Due to the high number of these negative selection mutations, we suggest that there was one (or more) mutations that caused a change in the fitness landscape, whereby many previously deleterious mutations became neutral/adaptive.
In parallel, and complementary to the above, @GuptaR_lab, @PeacockFlu and others showed, Omi might have changed its tropism away from lung cells, and this is (hopefully) in line with clinical data emerging.
So - SARS-CoV-2 might have made a large phenotypic change, and might have moved to a remote region in the fitness landscape as compared to previous variants. One open question remains – how far is Omi, if this is the case, from the peak in this new region?
This means we will need to start gathering new data, genomic and clinical. And we need to keep in mind that applying the knowledge we have on disease caused by previous variants will drive us to the wrong conclusions.
The data for this analysis is available here :
docs.google.com/spreadsheets/d…
docs.google.com/spreadsheets/d…
• • •
Missing some Tweet in this thread? You can try to
force a refresh