1. Update on the "Yeast Hypothesis" (Revised)
Differential enrichment of yeast DNA in SARS-CoV-2 and related genomes supports synthetic origin hypothesis [Version 3)
f1000research.com/articles/10-91…
Differential enrichment of yeast DNA in SARS-CoV-2 and related genomes supports synthetic origin hypothesis [Version 3)
f1000research.com/articles/10-91…
https://twitter.com/BillyBostickson/status/1408110651386466306
2. Amendments from Version 2
Furin cleavage site data and discussion included. Identical sequence stretch on S. cerevisiae chromosome XIII identified and suggested as potential recombination donor for the furin site insert in the SARS-CoV-2 spike glycoprotein
Furin cleavage site data and discussion included. Identical sequence stretch on S. cerevisiae chromosome XIII identified and suggested as potential recombination donor for the furin site insert in the SARS-CoV-2 spike glycoprotein

3. Revised version 3 includes new data regarding the origin of the unusual FCS in SARS-CoV-2. Indeed, with these new data & evidence, it suggests that this critical site is the product of a recombination event after synthetic passage in genetically manipulated S. cerevisiae cells
4. . Overall, the research presented in this article now provides strong evidence that SARS-CoV-2 has an origin in synthetic yeast, and as such is not a product of natural evolution alone.
5. Highlighted Revisions - 1
"At this junction, we detect a highly specific stretch of yeast DNA encoding for the critical furin cleavage site insert PRRA, which has not been seen in other lineage b betacoronaviruses"
"At this junction, we detect a highly specific stretch of yeast DNA encoding for the critical furin cleavage site insert PRRA, which has not been seen in other lineage b betacoronaviruses"
6. Highlighted Revisions - 2
"Our data specifically allow the identification of
the yeast S. cerevisiae as a potential recombination donor for the critical furin cleavage site in SARS-CoV-2"
"Our data specifically allow the identification of
the yeast S. cerevisiae as a potential recombination donor for the critical furin cleavage site in SARS-CoV-2"

7. Highlighted Revisions - 3
"Of special interest in this analysis was a 16 base sequence TTCTCCTCGGCGGGCA near P2 between position 23599 & 23614, which corresponded to the furin cleavage site & identically aligned with bases [810386..810401] from S. cerevisiae chromosome XIII"
"Of special interest in this analysis was a 16 base sequence TTCTCCTCGGCGGGCA near P2 between position 23599 & 23614, which corresponded to the furin cleavage site & identically aligned with bases [810386..810401] from S. cerevisiae chromosome XIII"

8. Highlighted Revisions - 4
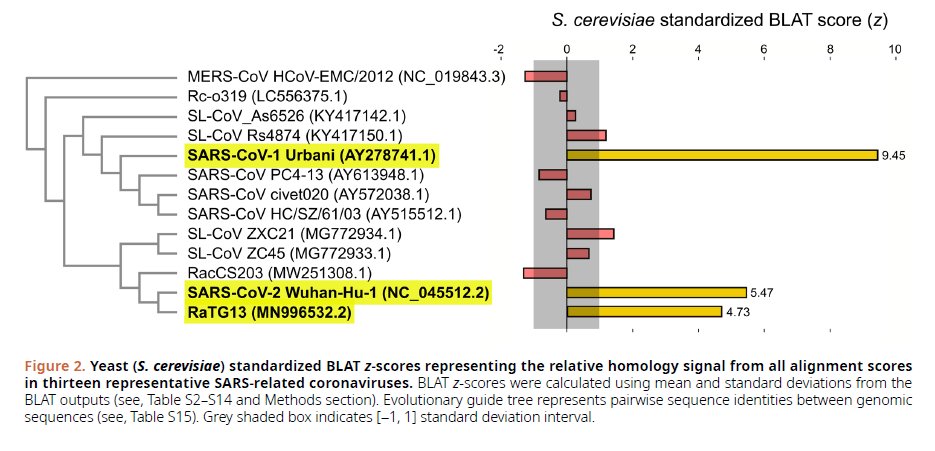
"Yeast (S. cerevisiae) standardized BLAT z-scores representing the relative homology signal from all alignment scores in 13 representative SARS-related coronaviruses"
Note that only, SARS-COV 1, SARS -COV 2 and RaTG13 show Yeast anomalies.
"Yeast (S. cerevisiae) standardized BLAT z-scores representing the relative homology signal from all alignment scores in 13 representative SARS-related coronaviruses"
Note that only, SARS-COV 1, SARS -COV 2 and RaTG13 show Yeast anomalies.
9. Highlighted Revisions - 5
"According to our data, this cleavage site is specifically compatible with a recombination event including chromosome XIII of S. cerevisiae, which shares a unique nucleotide sequence that encodes
the necessary insert PRRA"
"According to our data, this cleavage site is specifically compatible with a recombination event including chromosome XIII of S. cerevisiae, which shares a unique nucleotide sequence that encodes
the necessary insert PRRA"
10. Any questions?
Corresponding author: Andreas Martin Lisewski
a.lisewski@jacobs-university.de
Corresponding author: Andreas Martin Lisewski
a.lisewski@jacobs-university.de
• • •
Missing some Tweet in this thread? You can try to
force a refresh




