If you work w/ single-cell RNA-seq & are performing RNA velocity analyses, you might find this @GorinGennady et al. preprint w/ Meichen Fang & Tara Chari of interest. It's a deep dive into the method, and navigation of the 67 pages may be aided w/ this🧵1/
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
As a starting point, it's worth noting that the two popular packages right now, scVelo (@VolkerBergen et al. from @fabian_theis' lab) and velocyto (@GioeleLaManno et al. from the @slinnarsson and @KharchenkoLab labs), yield discordant results on a simple example (see below). 2/ 

The inferred directions should recapitulate a known differentiation trajectory from radial glia to mature neurons. However, scVelo reverss the trajectory, despite "generalizing" velocyto & relying on a better model. Also sometimes it's scVelo that works well. So what gives? 3/ 

Getting to the bottom of what RNA velocity software is really doing and how implementations relate to theory is a project that has its roots in @GorinGennady's rotation, where working with @vallens he published "protein acceleration". 4/ genomebiology.biomedcentral.com/articles/10.11…
@GorinGennady was unsettled by the heuristics in the workflows, and the strong, biophysically unrealistic, assumptions underlying the models. Arriving at an understanding of what is going on was a monumental task, whose scope is clear considering what a workflow constitutes: 5/ 
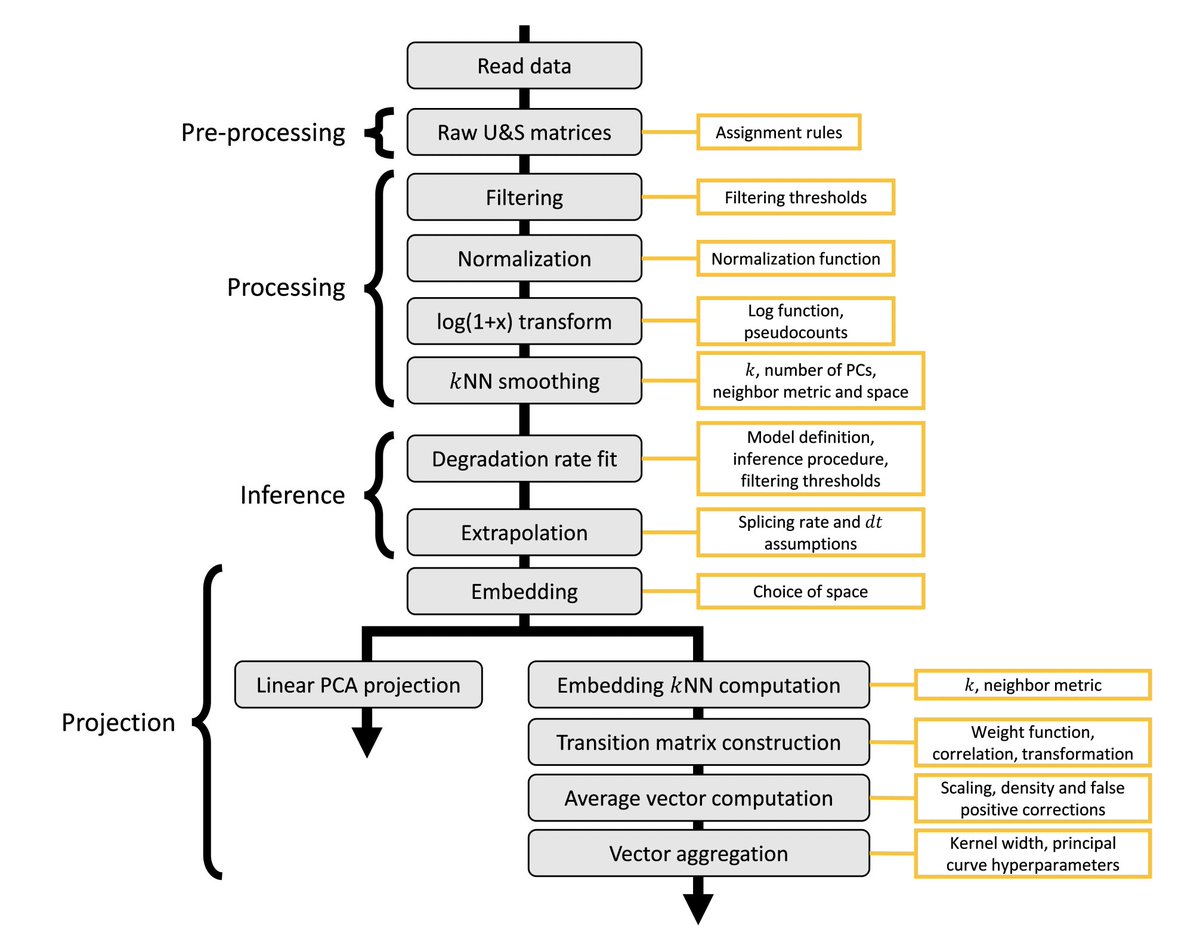
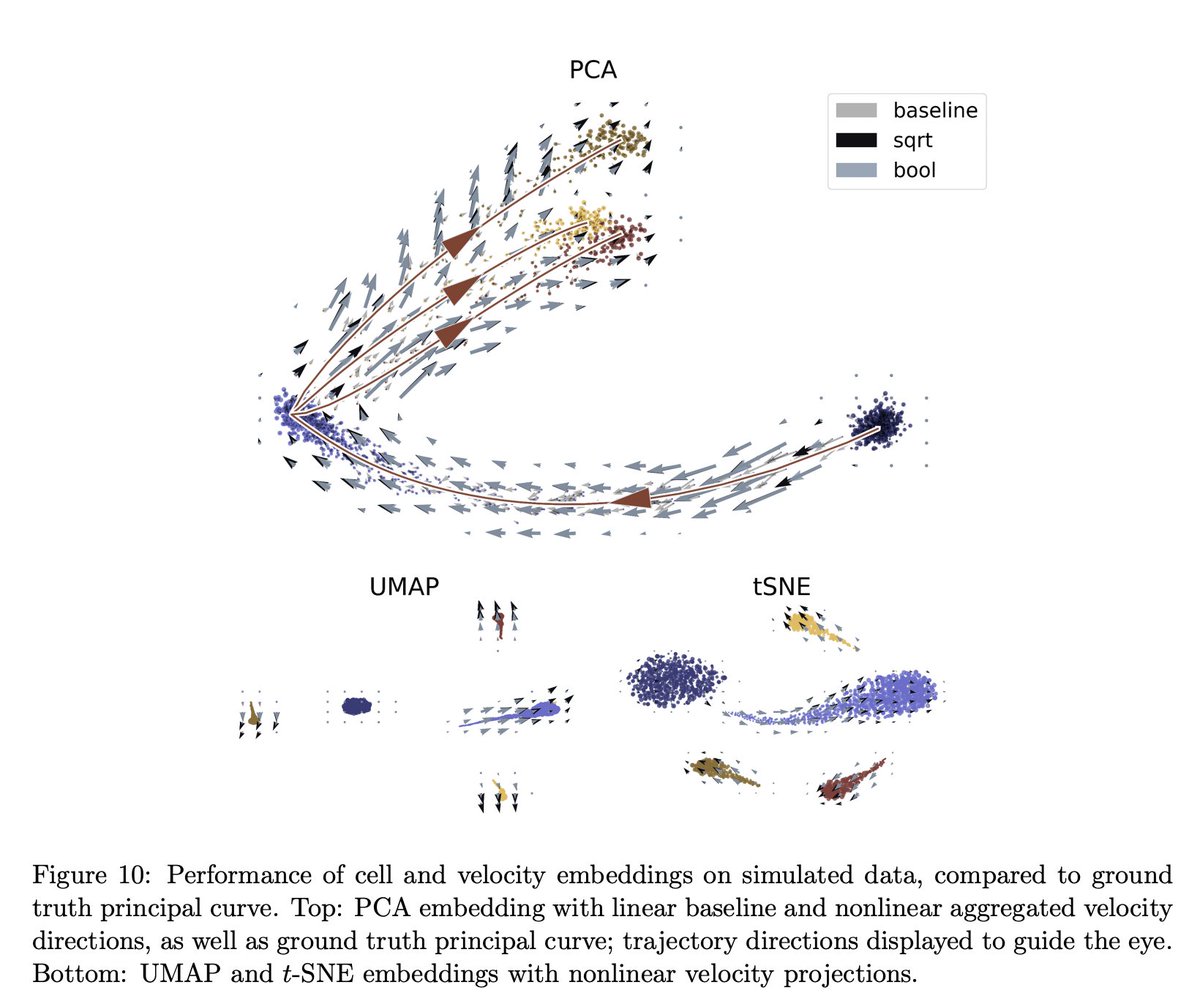
To get a handle on RNA velocity, we started by examining how different choices, for the different steps, affect performance. There is a lot to go through, and results are provided in the section "Logic and Methodology". 6/ 

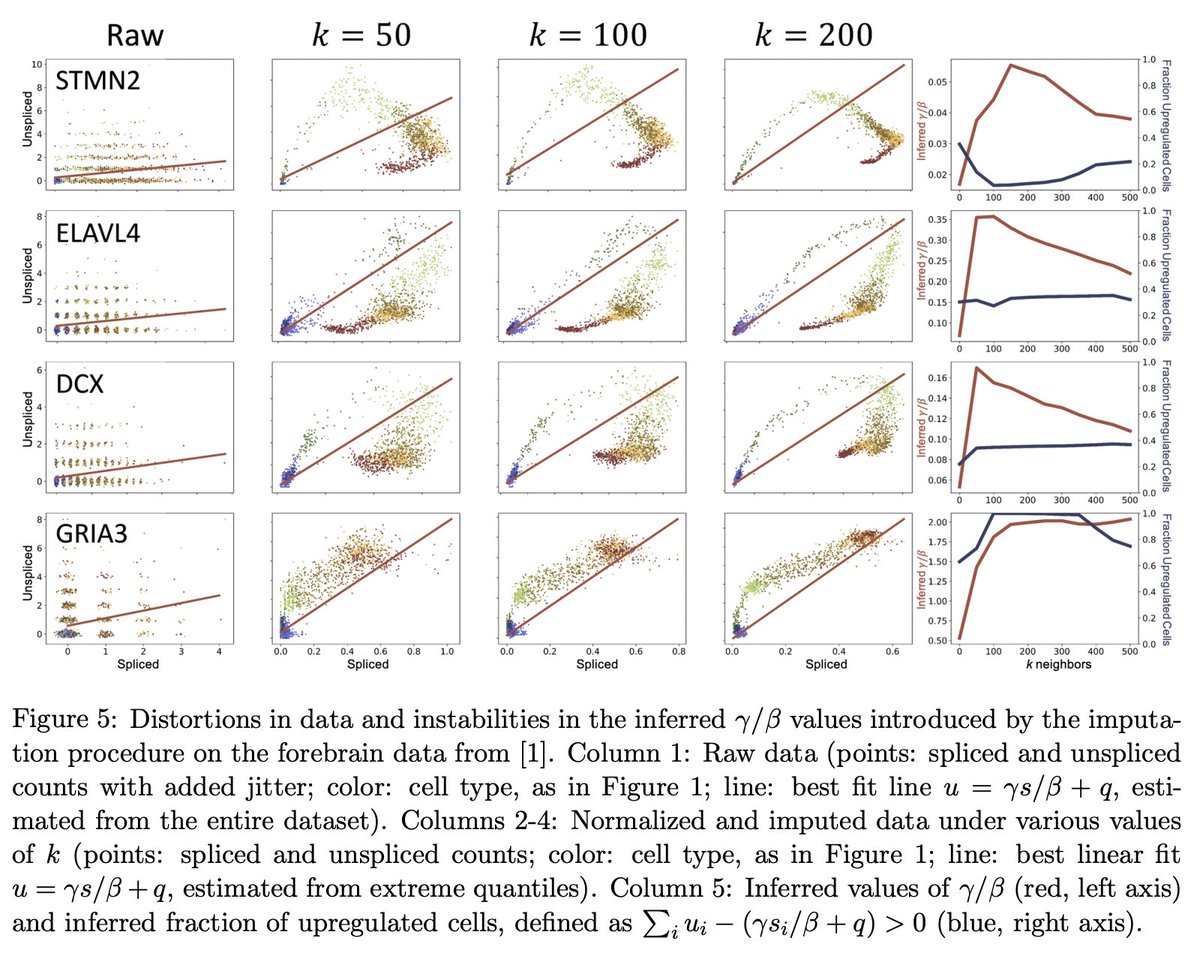
For example, curious how the amount of smoothing, i.e., choice of number of neighbors (in the k-nearest-neighbor graph construction) affects results? See Figure 5. 7/ 
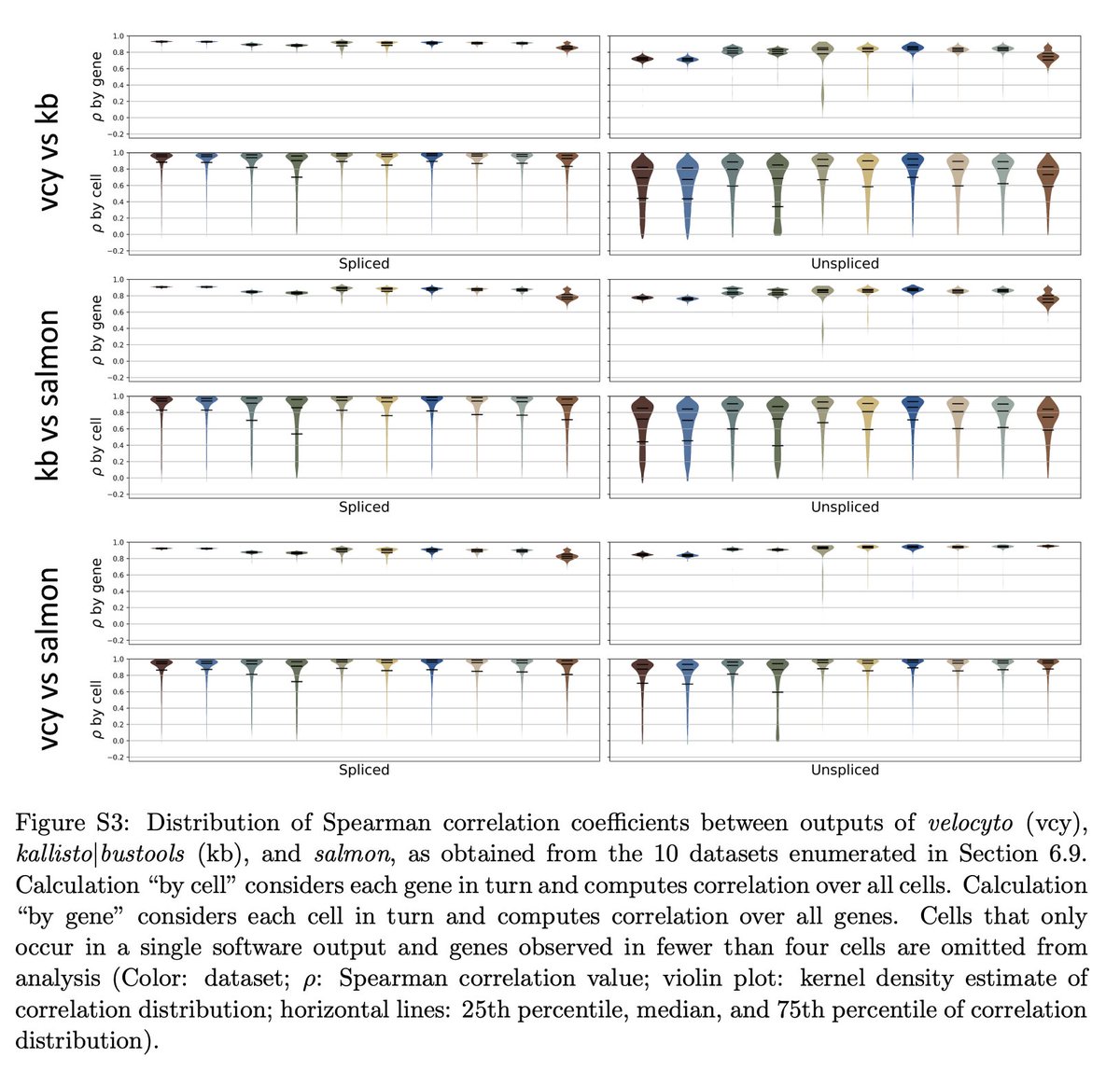
Interested in how the choice of pre-processing workflow affects results? See Supplementary Figure 3 (this question alone was studied by @CSoneson et al. in journals.plos.org/ploscompbiol/a… but we extended the number of datasets examined). 8/ 
But perhaps most important, is the choice of model, and the assumptions that go into it. The schema looks simple (below), but it obscures significant complexity. 9/ 

Consider, for example, just scVelo and velocyto. The following models (used in different publications), all make different assumptions about the data-generating processes: 10/ 
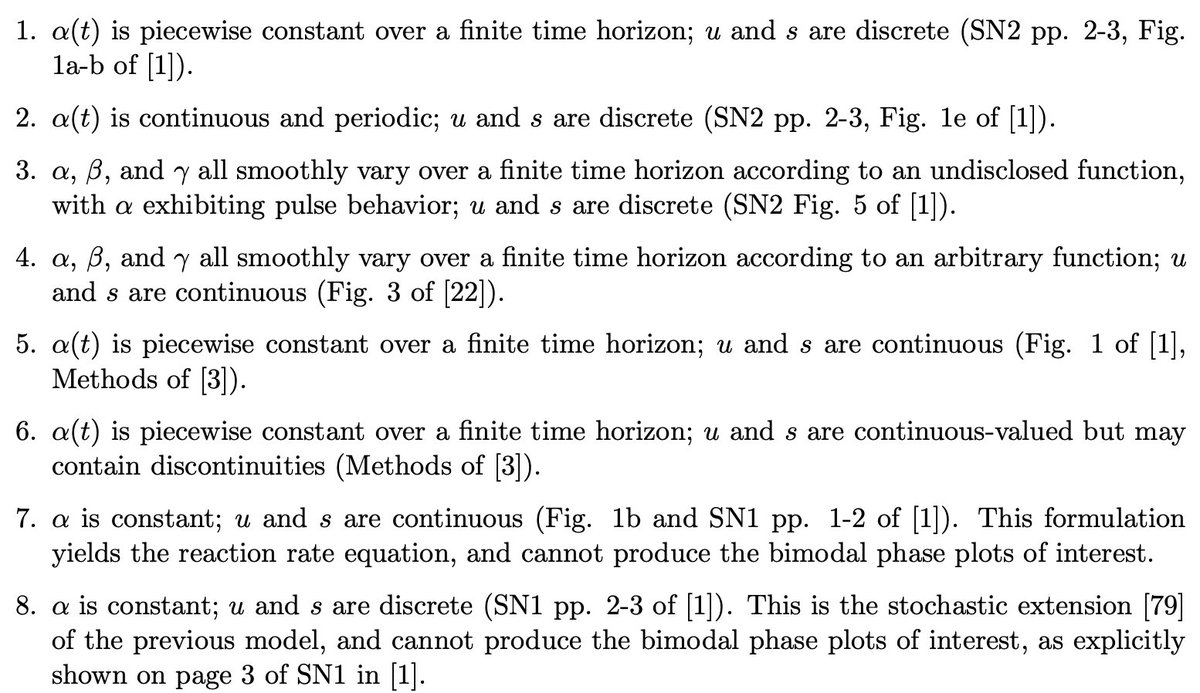
We examine the implications of these assumptions in detail for velocyto and scVelo in the paper, although our results are relevant for other recent extensions of them as well, e.g. CellRank from the @dana_peer and @fabian_theis labs, dynamo from the @JswLab lab, etc. 11/
What does it mean to understand the modeling assumptions for RNA velocity? Ultimately, it requires developing a coherent framework from which existing methods can be understood as special cases. That is what we do in Section 4. The key is a missing piece that we elucidate... 12/
Specifically, we highlight the importance of treating occupation measures as part of the model explicitly. What this means is that while cells are sampled simultaneously in #scRNAseq experiments, it is crucial to model their underlying process time via a sampling distribution.13/
The resultant combinatorial optimization that must be solved for RNA velocity is difficult. We explain in the manuscript how the problem is related to that of (pseudotime) trajectory inference, & how heuristics for that problem such as traveling salesman problem, are related. 14/
By revisiting the velocyto and scVelo models from this general vantage point, the nature of their assumptions, and the implications thereof, become clear (see Sections 4.7 and 4.8) . 15/
Without a rigorous model foundation, RNA velocity is not very meaningful, but that hasn't stopped people from using it to make pretty figures with arrows suggesting cell trajectories. We looked at the embeddings in detail, and that forms another large chunk of the paper. 16/
We also ran a negative control, which highlights that absence of absence does not imply presence! 18/ 

There is a lot more in the paper, than what I've highlighted here. We went carefully through every step of RNA velocity workflows, including some I haven't discussed in the thread (e.g. normalization). 19/
One interesting analysis we did is to examine what happens in a "binarized" analysis. Essentially, instead of computing transition probabilities based on the velocity values, we computed them based on signs. It reveals something about what RNA velocity is *really* showing. 20/ 
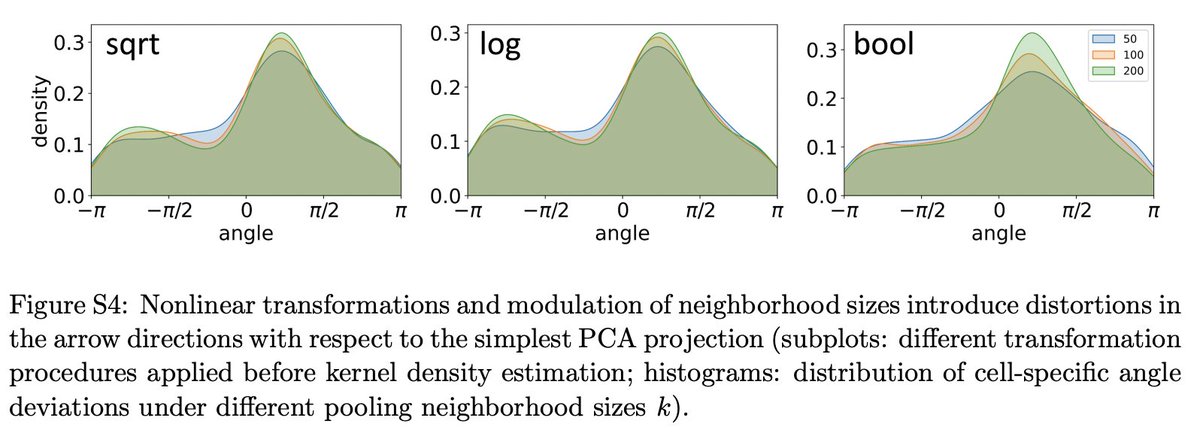
The bottom line is this: RNA velocity is the product of two (largely) independent scientific threads: one is fluorescent transcriptomics, in which single-molecule fluorescence technologies motivated the development of interpretable stochastic models of biological variation. 21/
This lineage has led to work such as elifesciences.org/articles/69324 by Ham et al. from the @theosysbio lab. 22/
On the other hand, sequence census methods (nature.com/articles/nmeth…), such as single-cell RNA-seq, leverage high-throughput sequencing to produce large datasets, and are typically analyzed from a phenomenological perspective. 23/
Whereas the goal with sequence census assays is to remove noise, from a modeling perspective noise is the biophysical phenomenon of interest! Intrinsic and extrinsic noise in @ElowitzLab et al.'s single-cell classic science.org/doi/full/10.11… are the result, not the nuisance. 24/
In summary, RNA velocity as currently practiced is problematic for many reasons, but there is balm in Gilead. @AmitZeisel et al.'s work (embopress.org/doi/full/10.10…) that forged the path towards RNA velocity has an interesting legacy that is only starting to be explored. 25/
This project has been several years in the making. In addition to @GorinGennady's work, Meichen Fang & Tara Chari implemented many of the experiments. I don't like to throw around the phrase "tour de force", but this project was truly that. 26/
As is now standard for the work from our lab, all of the code to reproduce the results and figures in the paper is available via @GoogleColab notebooks at github.com/pachterlab/GFC… 27/27
• • •
Missing some Tweet in this thread? You can try to
force a refresh