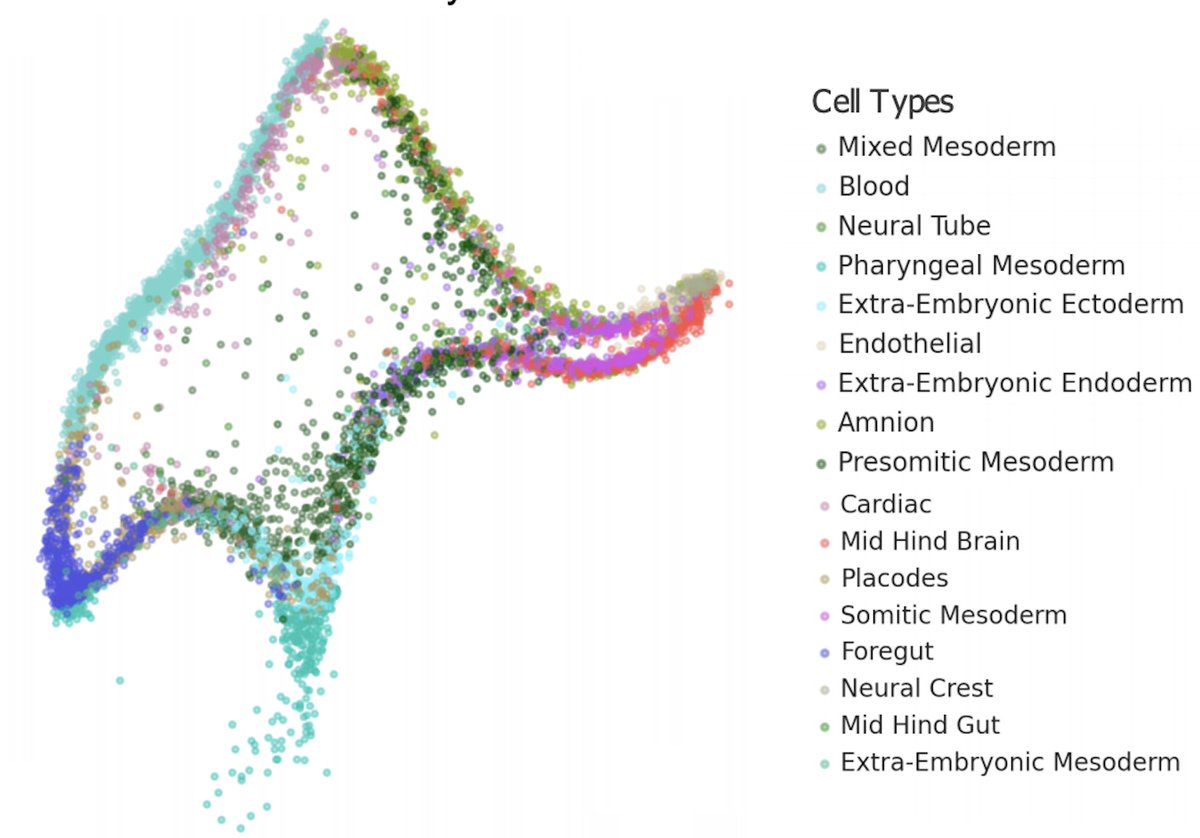
The 17 #BICCN @nature papers on the primary motor cortex in mouse (+some human & marmoset) that were published yesterday are a major step forward in terms of open science for an @NIH consortium. For reference, links to the open access papers are here: nature.com/collections/ci… 1/🧵
First, the #BICCN required preprints of all the papers to be posted on @biorxivpreprint, and as a result the papers were already online 1-1.5 years ago. Of course the final versions now published have been revised in response to peer review. 2/
Speaking of peer review, almost all the papers were published along with the reviews. In combination with the preprints, this provides an unprecedented view of how consortium work is reviewed and how authors respond. Real data for this perennial debate:
https://twitter.com/arjunrajlab/status/1444684256895283201?s=203/
Some of the reviews were superficial. For example referee #1 of the “flagship” paper (nature.com/articles/s4158…) wrote one paragraph summarizing the work + 3 minor comments (for a paper whose goal was to synthesize results from complex data published in 11 other papers!). 4/ 



Some reviews were brutally honest. Referee #1 of nature.com/articles/s4158… wrote "what we have..is a very well collected catalogue utterly devoid of either a conceptual framework or even an idea... the experience is like reading a phone book" . They signed the review (@blamlab).5/
Some reviews were serious(ly helpful). E.g., in our paper (one of the 17, namely @sinabooeshaghi et al., nature.com/articles/s4158…), a referee was concerned about batch effects & artifacts, leading us into a deep dive that revealed batch effects in the consortium #scRNAseq data. 6/
This is not the first time reviews of papers are published (@eLife has been doing this for a while), but having the referee reports (+ responses & non-responses) exposed for an entire consortium-worth of papers is a dataset ripe for study (& beyond the scope of this thread). 8/
BTW, not all labs agreed to publication of their referee reports. 6 papers out of 17 opted out:
nature.com/articles/s4158…
nature.com/articles/s4158…
nature.com/articles/s4158…
nature.com/articles/s4158…
nature.com/articles/s4159…
nature.com/articles/s4146…
9/
nature.com/articles/s4158…
nature.com/articles/s4158…
nature.com/articles/s4158…
nature.com/articles/s4158…
nature.com/articles/s4159…
nature.com/articles/s4146…
9/
Another aspect of open science is freely available data. In that regard, the #BICCN consortium has been exemplary. All the data generated is freely available, for example the #scRNAseq data used in @booeshaghi et al. is here (and has been for years): data.nemoarchive.org/biccn/grant/u1… 10/
However, as pointed out in a recent paper by @autobencoder, @michaelhoffman, @markowetzlab, @suinleelab, @GreeneScientist, and @stephaniehicks, data is not enough. Models and code are also essential and therefore a key part of open science. 11/ nature.com/articles/s4159…
Unfortunately, despite the fact that computational methods (including machine learning tools) are an essential piece of the #BICCN, many of the consortium papers fail to even medal by the standards of @autobencoder et al.
https://twitter.com/michaelhoffman/status/1441198676706091015?s=20. 12/
Many papers released no code to reproduce results or figures from their papers, and omitted key analysis details. This is not specific to #BICCN; it reflects widespread belief in the genomics community that data trumps methods, and rejection of the idea that #methodsmatter. 13/
However, most of the data generated for the 17 @nature papers published by the #BICCN was generated quickly and its the analysis that has taken several years. The difficulty of analysis can be seen in the papers that did release code (some achieved bronze 🥉). 14/
See., e.g. github.com/AllenInstitute… from Bakken et al. (nature.com/articles/s4158…) that shows just how challenging it is to perform analysis of the #BICCN data (and how useful it is to have the code). 15/
The #BICCN datasets were so large that it was challenging to enable reproducibility. In our paper (nature.com/articles/s4158…) @booeshaghi struggled to achieve "one-click" reproducibility with @GoogleColab that we strive for. I'd say we achieved bronze trending towards silver.. 16/
In summary, the steps taken towards open science by the #BICCN represent real progress. Having now participated in consortia from the mouse genome (2002) to the mouse brain (2021), I can say the progress is astounding. But we're still not at platinum.17/17
https://twitter.com/lpachter/status/1441257875225989125?s=20
• • •
Missing some Tweet in this thread? You can try to
force a refresh