
Thrilled to announce my 1st paper from @stevens1lab is now published @NatureNeuro. “Dissection of Artifactual and Confounding Glial Signatures by scRNA-seq of Mouse and Human Brain”. Why is it important? a 🧵👇 #singlecell #microglia #methodsmatter 1/n
nature.com/articles/s4159…
nature.com/articles/s4159…
While the title frames this work in terms of brain, a KEY takeaway from new data in this version is that this is broadly applicable across basically all scRNA-seq (and RNA-seq) studies (especially in immunology)... 2/n
...and the artifact we discuss is unfortunately highly prevalent in current literature. So stick around even if brain isn’t your thing 😉. We thoroughly characterize the issue and provide a robust flexible solution to eliminate it as well. 3/n
Next, I need to give HUGE thanks to everyone involved. This paper & arduous revision process truly would not have been possible without all of your work and support!! 4/n
Tagging here. Truly a huge collab effort.
@AlecJWalker, @tkam80, @thammondglia, Lasse Dissing-Olesen, Sarah Murphy, Adam Young, Robin Franklin, @danjgaffney, @YvankadeSoysa, @soyonhonglab, @ConnorDufort, @Abdul_Squared, @LaLilli11, @DavidHaflerMD, and everyone not on Twitter. 5/n
@AlecJWalker, @tkam80, @thammondglia, Lasse Dissing-Olesen, Sarah Murphy, Adam Young, Robin Franklin, @danjgaffney, @YvankadeSoysa, @soyonhonglab, @ConnorDufort, @Abdul_Squared, @LaLilli11, @DavidHaflerMD, and everyone not on Twitter. 5/n
And last but not least big thanks to my mentor @stevens1lab for believing in paper and fighting with us during tough revisions. 6/n
With that done let’s dig in!!
First off it’s been a long road in review (sub. Dec 2020) so you can catch up on what was in preprint via thread I wrote back then:
7/n
First off it’s been a long road in review (sub. Dec 2020) so you can catch up on what was in preprint via thread I wrote back then:
https://twitter.com/samuel_marsh/status/1334646157994303488?s=21
7/n
One final preface. The final results/reanalysis we perform does require a lot of context. Way more than twitter & more than the journal allows (9 SI Notes/Discussion). I promise it's good read and important & encourage everyone to check it out to understand things completely. 8/n
Astute among you may be looking at things and saying “Hey aren’t the main figures the same as preprint?” Yes! That is because the main conclusions and the data supporting them were solid and the main focus. But new text and supplement is where things really get good 😃
9/n
9/n
Among the largest changes were significant updates to text and SI figures to frame the larger issue of ex vivo induced transcriptional changes, their prevalence, their impact on downstream analysis, and the applicability of our experimental solution beyond the brain 10/n
While we had described the prevalence of this issue in microglia field a bit, we now do so more in depth and begin to address two important contexts: atlas studies and case/control (comparative) studies and how the exAM signature causes issues with both types of studies. 11/n
Atlas/survey studies are invaluable resources for field. They are used to set benchmark for what cells are present, how they develop/change, and more and more are used as reference datasets to align and annotate future studies. 12/n
However, as we note many of these studies also contain the exAM signature in microglia which results in over-annotation of microglial heterogeneity. I'm not here to call out any studies but there is significant need to raise awareness of this signature so that these... 13/n
... incorrect annotations are not used in the future or as starting points for new studies. 14/n
In addition to atlas studies, the number of case/control studies using scRNA-seq are increasing rapidly. So we wanted to examine how the presence of exAM signature might cause issues for downstream analysis of these kinds of studies. 15/n
One example is from Keren-Shaul et al., 2017 where the authors noted that one of their datasets was isolated using different protocol and had a drastic increase in genes that are part of exAM signature. This provided us with nice case-study to understand the impact of exAM. 16/n
We reanalyzed just this dataset with an eye on the exAM signature. We find is that the exAM signature is higher is cells from WT mice or from non-"DAM" cells in AD mice. We believe that this reflects the fact that microglia already polarized towards one response in vivo... 17/n 

...are slower to change & respond to different stimuli (ex vivo dissociation). Example highlights two important things. 1. This differential presence of exAM can lead to misidentification of DEG if not accounted for. 2. Computational strategies are currently problematic... 18/n
Regressing the signature during clustering or removal of cells high in exAM signature (commonly performed in literature) differentially effects cells from different genotypes and adds additional bias to the downstream results. Hence we currently advise against these methods 19/n
Another common comparative study that has become very popular is comparison of analogous cell populations across tissue compartments. We highlight how the ex vivo artifact can cause significant issues here too. 20/n
We show 2 examples of studies comparing immune populations in the blood vs tissue-resident (in mice & humans). We demonstrate that differential tissue processing (undigested blood vs digested tissue) leads to aspects of exAM signature being characterized as tissue-resident. 21/n 


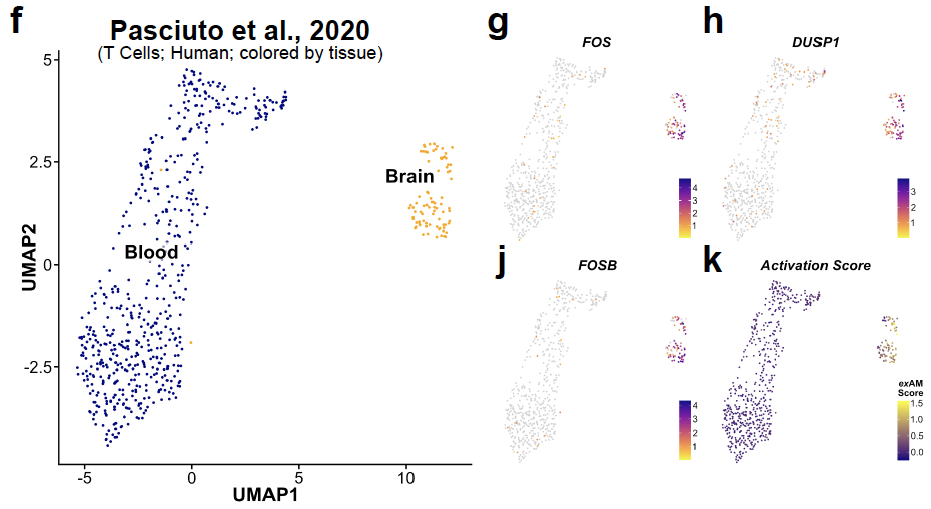
To further examine this we collaborated with David Hafler and @LaLilli11. They performed experiment to "digest" human PBMCs with or without or inhibitor cocktail present. 22/n
What we find is that when when conditions analogous to tissue digestion are applied PBMCs dramatically alter their transcriptomes and express many of the exAM markers previously attributed to tissue-resident signatures.
23/n
23/n

This is also a significant issue for cell surface proteomics flow, CyTOF, CITE-Seq as comparing digested and undigested tissues via those techniques will yield differences that are simply the result of extracellular receptor cleavage by enzymes.
24/n
24/n

We have couple of other new experiments/figures. First we verify that the extra time of droplet generation from 10X V3.0 (8 min) > V3.1 (17 min) does not induce exAM signature. 25/n 

We also more deeply examine the expression of a few genes in exAM signature including genes previously implicated as key components of microglia to demonstrate that there is virtually no expression outside of exAM clusters. 26/n 

Finally, we added a lot more discussion of the similar signature that we find in post-mortem human tissue. 27/n
Especially as it concerns acute pre-mortem factors such as agonal state, cause of death, etc and the potential for those factors to be collinear with other common meta data (e.g., Age, RIN, etc) 28/n
We make the case in the discussion but I'll also do so here that meta data from human snRNA-seq studies is CRITICAL & we need to aspire to share as much as we possibly can. This is not overnight change but to truly dissect some of the large sources of variability in human... 29/n
...data we need a lot more meta data than is usually provided. Great analyses in recent paper from @towfiqueRaj @lopeskp nature.com/articles/s4158… as well as one from Allen Institute (Miller et al., 2017) clearly demonstrate the value in meta data. 30/n elifesciences.org/articles/31126…
I truly believe this is critically important issue for the scRNA-seq field to address because the signature is way to common in lit. Our experimental solution & inhibitor cocktail provides a robust & flexible method to avoid ex vivo issues in your data. 31/n
Another plug I will always make from now on is open data and share as much as possible (FYI my methods section has higher word count than the actual manuscript) but I hope that makes it easier for anyone reading to follow.
32/n
32/n
All of the code needed to reproduce the scRNA-seq/snRNA-seq analysis objects for new data in the manuscript and my reanalysis of previously published studies can be found at GitHub repo:
github.com/samuel-marsh/M…
33/n
github.com/samuel-marsh/M…
33/n
All of the raw AND processed sequencing data (fastq/BAM and matrix files) can be found via links/accession numbers in the article and via links at GitHub code repo. (Some of it is controlled access, beyond my control).
34/n
34/n
That's all for now but there's more in the paper and like I mentioned even more in the SI Figures and SI Notes. If you have questions feel free to reach out! If you are still here thanks for reading and I hope this helps you in your future scRNA-seq studies/analysis!! 35/35
Oh my goodness so embarrassed I missed tagging @macosko here! Big thanks of course!!! This wouldn’t have been possible without your help, guidance and collaboration! Sorry!!!
• • •
Missing some Tweet in this thread? You can try to
force a refresh



