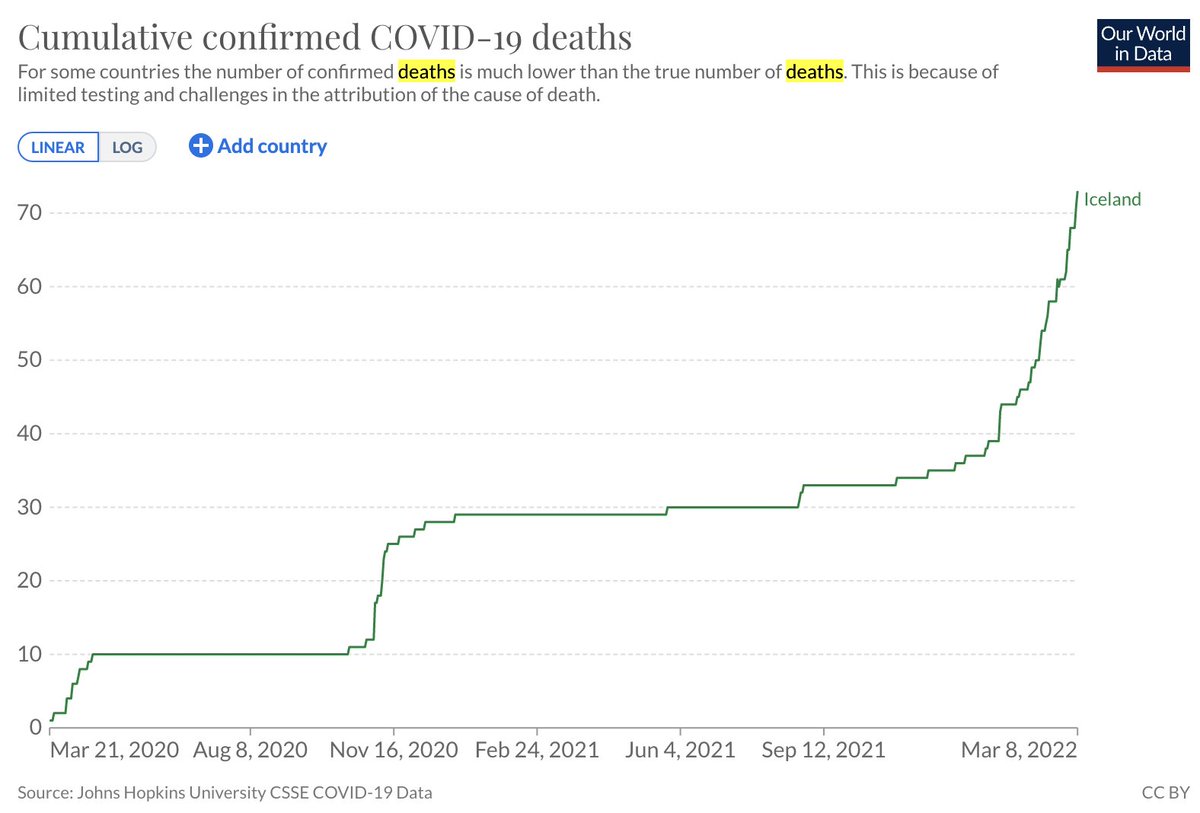
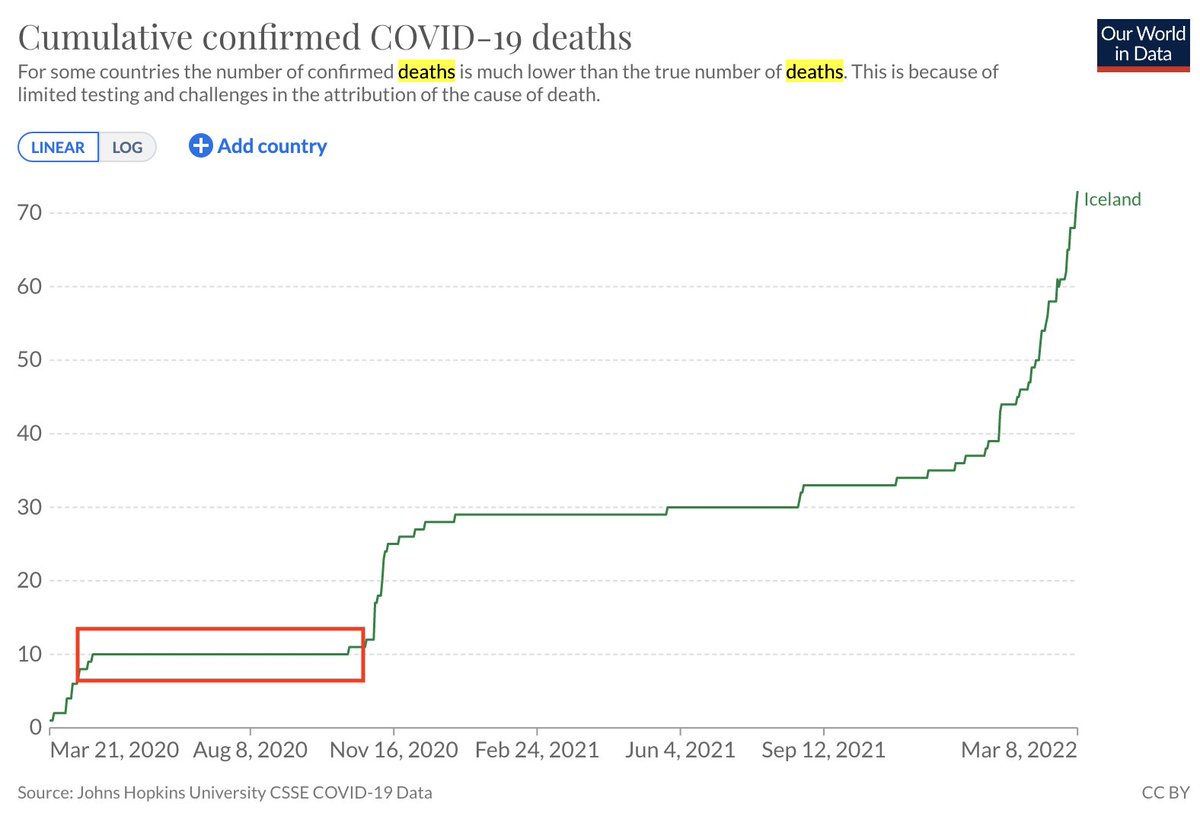
Analysis of #scRNAseq requires constant, tedious, interaction with genomics databases. To facilitate querying from @ensembl et al., @NeuroLuebbert developed gget:
biorxiv.org/content/10.110… (code @ github.com/pachterlab/gget).
gget has many uses; a 🧵on the its amazing versatility: 1/
biorxiv.org/content/10.110… (code @ github.com/pachterlab/gget).
gget has many uses; a 🧵on the its amazing versatility: 1/
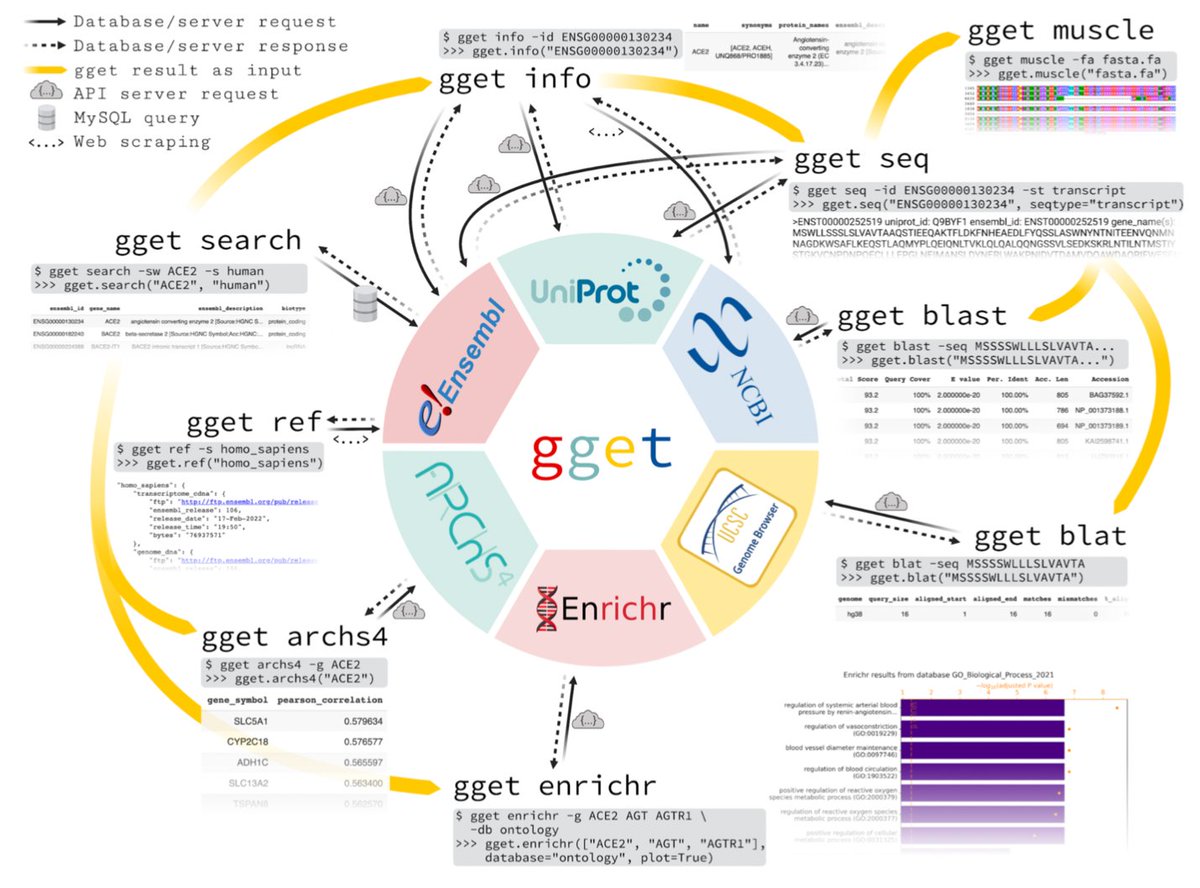
gget works from the command line or python. Just `pip install gget`.
Need reference files for your analysis? 2/
Need reference files for your analysis? 2/
https://twitter.com/ensembl/status/1149633319933374464
Gene ID conversions *used* to be a pain... 4/
https://twitter.com/tina_kesha/status/1463281367186296833
Here's a common task that anyone who has done #RNAseq or #scRNAseq has faced: 6/
https://twitter.com/jomcinerney/status/1516383914905980932
This tasks requires a lot of clicks on the @GenomeBrowser... 8/
https://twitter.com/ines_boehm/status/1516097766413455365
You don't need to take a class to BLAST with gget: 10/
https://twitter.com/MaidenLab/status/1219671030270255106
And no more need to choose sides in BLAT vs. BLAST twitter wars... 12/
https://twitter.com/nilshomer/status/1505241083151740932?s=20&t=KCTfCGJWoBJuPnnolg30kQ
We found in our work that aligning genes and/or isoforms can be useful in the course of #scRNAseq analyses. So we built an alignment interface. 14/
https://twitter.com/ScottESolomon/status/1202315983895048192
This is an excellent question... 16/
https://twitter.com/Caroline_Bartma/status/1024085429149204486
gget provides a solution via a convenient interface to @alexlachmann, @AviMaayan et al.'s ARCHS4. 17/ 

Hopefully gget also helps in training / classes / workshops. 18/
https://twitter.com/hostmicrobe/status/1124088753419628544?s=20&t=rH4zl7uz0AP7dmAc50fGfA
Now... every PI wants a GO enrichment analysis. 19/
https://twitter.com/chelysheva_i/status/1411629899333386247
We got that covered (thanks to Enrichr from @MaayanLab). 20/ 

Putting it all together, gget can, in a few lines, easily take care of more complicated tasks such as this one... 21/
https://twitter.com/anthony_berndt/status/1446304325702090755
... just 5 calls to gget!
The ability to do free-form searching is, we think, going to be particularly useful. 22/
The ability to do free-form searching is, we think, going to be particularly useful. 22/

We'd appreciate any feedback via Github, including requests for additional features: github.com/pachterlab/gget 23/
Finally, gget was written entirely by @NeuroLuebbert, but thanks to @agalvezmerchan, @kreldjarn, Matteo Guareschi, @sinabooeshaghi, and @lioscro for helpful feedback and advice on the tool. 24/25
Special thanks to Dash Coffee Bar for facilitating deep contemplation of the RNA-seq smoothie metaphor...
25/25
25/25

• • •
Missing some Tweet in this thread? You can try to
force a refresh