Okay, so here are my comments on the new paper in @Nature announcing palaeogenetic identification of the origin of the Black Death. Since this will be a long thread, a few general pointers. 1/n
Terminology:
- BD = Black Death
- Spyrou (unless otherwise specified) = Maria A. Spyrou et al. 2022 study in @Nature (nature.com/articles/s4158…)
- SNP = single nucleotide polymorphism (to be explained below)
- phylogenetics = study of evolution at the genomic level
2/n
- BD = Black Death
- Spyrou (unless otherwise specified) = Maria A. Spyrou et al. 2022 study in @Nature (nature.com/articles/s4158…)
- SNP = single nucleotide polymorphism (to be explained below)
- phylogenetics = study of evolution at the genomic level
2/n
Audience: I'm writing with fellow historians & medievalists uppermost in my mind. Many #twitterstorians & #MedievalTwitter followers know a little bit about the transformations in plague history in the past decade. But not everybody has time to keep up on all the details. 3/n
This thread demonstrates what's at stake in the claims being made, & the methods being used, in this new study in @Nature. And that is why I am also addressing this 🧵 to the editors at Nature, who did not solicit peer review from historians. I aim to show why that matters. 4/n
A long 🧵 so here is its basic structure:
Part I: some background on plague research & why palaeogenetics (= aDNA) has been transformative
Part II: this study: what we already knew, what's new
Part III: "The Case of the Missing SNP"
Part IV: "The Case of the Missing Genomes"
5/n
Part I: some background on plague research & why palaeogenetics (= aDNA) has been transformative
Part II: this study: what we already knew, what's new
Part III: "The Case of the Missing SNP"
Part IV: "The Case of the Missing Genomes"
5/n
Part I: so, what's been going on in Plague Research? Lots. And I've been talking about it for years. My interest in better narratives of plague is 2-fold: 1) we historians have to teach this in our classrooms. The narratives that've been available have been lousy. Full stop.
6/n
6/n
2) Plague is the best "model organism" for studying pandemics. We know more about its history now than for any other major infectious disease. Yes, it's (mostly) a controlled disease now, not a threat. But the human aspects of pandemics still need to be understood.
7/n
7/n
Here's a ready-made thread (pub'd in Oct 2020) w/ a summary of the highlights in the genetics of plague (#YersiniaPestis) as it relates to its evolutionary history:
8/n
https://twitter.com/monicaMedHist/status/1317813805703856134.
8/n
And here's a teaching guide, for those who need to craft a narrative about the BD (see beginning of thread for abbreviations) in their classrooms: academia.edu/65148743/Green….
9/n
9/n
And here's a general bibliography on the #2ndPlaguePandemic, which is actually a better term now for the BD because the latter has been framed, in the historiography, too narrowly. docs.google.com/document/d/1x0…
10/n
10/n
One final point necessary as background: why has every newspaper in the world been carrying this story of 1 scientific discovery from 1 small village in Kyrgyzstan that you've never heard of? That's the science-publishing-industrial-complex at work:
11/n
https://twitter.com/monicaMedHist/status/1229826171270922241?s=20&t=8NPDGHw3H5vib3THb1ftXg
11/n
Okay, that was Part I. Part II looks at what is confirmatory about this @Nature study (i.e., confirming that what we thought we already knew seems to be right) & what is truly new (something we never knew before).
12/n
12/n
A truism of History: we're almost always building on the work of predecessors, rarely inventing (or "discovering") from scratch. For these 2 14thC Christian communities in what is modern-day Kyrgyzstan, researchers built on archaeological work done in the 1880s & '90s.
13/n
13/n
The history of Christian communities in Central & East Asia in the Middle Ages is absolutely fascinating. For an up-to-date survey of the evidence, I highly recommend this study by Mark Dickens: academia.edu/42671100/Syria….
14/n
14/n
So, we've known about this burial site for about 140 years. We've known about the inscribed gravestones, which in many instances indicate year of death. Also, name of the diseased, so in many instances you can know gender. And 10 gravestones indicated cause of death.
15/n
15/n
In all 10 instances it's the same: mawtana. Pestilence. But here's the thing: "pestilence" isn't a specific medical diagnosis. The editor of the archaeological report, Chwolson, thought it was "Pest." Plague. So did Robert Pollitzer, author of the 1951 @WHO manual on plague. 16/n
And that's how the story of Issyk Kul got into plague historiography: b/c Pollitzer *assumed* it was plague & b/c he *assumed* that the plague found in Central Asia must be connected to the plague that struck Europe in the middle of the 14thC: the BD. Was he right?
17/n
17/n
No way to prove it. The bones & the gravestones of these Christian communities had been scattered by the time Pollitzer was writing in the 1950s. And there was no way at the time to confirm one way or another whether plague (#YersiniaPestis) was involved in their deaths.
18/n
18/n
Now you can see what's novel about the @Nature study: 1) the researchers tracked down the scattered bones & gravestones (or as many as they could--some have been destroyed); & 2) they applied a technique for detecting pathogens that Pollitzer never could have dreamed of.
19/n
19/n
That technique is aDNA analysis (= palaeogenetics). Here's a good explainer, a video of a lecture that lead author Johannes Krause gave several years ago when explaining their retrieval of the pathogen that caused mass mortality in 16thC Mexico: .
20/n
20/n
The fact that the present study comes out of the lab that's been principally involved in a number of pathogen aDNA retrievals is important background to the comments that follow in Parts III & IV of this thread. That is, that these are the scientists leading this new field. 21/n
If, as I will argue, the best practitioners in the field--producing (I think it fair to assume) the best quality data possible from the material remains at hand--if even they cannot lay out substantive evidence to support the claims being made, what are we historians to do?
22/n
22/n
We'll return to that question later. In terms of summarizing what's new in the study, it would help to break down the questions into component parts:
1) what killed the people who died suddenly in 1338-39 of "pestilence"? A: now we can say definitely that is was plague. 23/n
1) what killed the people who died suddenly in 1338-39 of "pestilence"? A: now we can say definitely that is was plague. 23/n
This confirms Chwolson's speculations from the 1880s, & Pollitzer's from 1951. Yeah! In fact, Many of the retrievals of #YersiniaPestis genomes in the past 10 years have been from sites where documentary evidence had already suggested plague's presence.
24/n
24/n
2) What was happening w/ #YersiniaPestis to cause this marmot-derived disease to enter into human bodies? Was it a change in the pathogen? Or humans? There's more that this important @Nature study accomplishes, but we'll get into that in Parts III & IV of this 🧵. More soon.
25/n
25/n
Part III: "The Case of the Missing SNP"
As I said, my main audience for this thread is fellow historians. And it is our occupational characteristic that we care about dates. (The kind on the calendar: not the ones you eat, or where you eat w/ a friend.)
26/n
As I said, my main audience for this thread is fellow historians. And it is our occupational characteristic that we care about dates. (The kind on the calendar: not the ones you eat, or where you eat w/ a friend.)
26/n
And any historian who has read the @Nature study w/ attentiveness saw that the authors accomplished a very slick move. They "disappeared" a whole century out from an emerging narrative about the #2ndPlaguePandemic. Part III looks at how they performed such prestidigitation.
27/n
27/n
Here's what I call a "SNP Bingo Card." I prepared this back in April. I've actually been tracking SNPs in the #YersiniaPestis phylogeny since 2014, when I edited a collection of essays on how historians & anthropologists might need to adjust their thinking about plague. 28/n 

But the real "light bulb moment" in my thinking about SNPs came in 2016, when, together w/ a biologist, I starting realizing why SNPs were so important in tracking plague's movements across historical landscapes: academia.edu/26543022/Tiny_…
29/n
29/n
Ever since then, when a new plague aDNA study comes out, I look at the SNP file in the accompanying data to confirm which SNPs (& how many) are characteristic of different Y. pestis lineages. So, what is a SNP? A mutation. We've all probably heard a lot about SNPs lately.
30/n
30/n
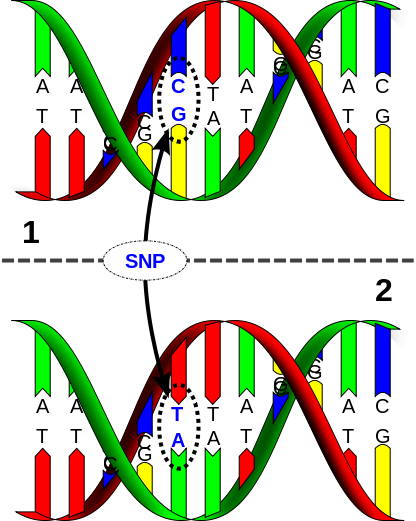
As I said, we historians care a lot about dating. "Change over time. Change over time." That's what we drum into the heads of our students. And dates are the anchors in any change-over-time analysis. But even though there are a gazillion calendrical systems in the world, ...
31/n
31/n
... not all the historical materials we work w/ are dated. So, one of the main skills we learn as historians is inferential chronological thinking: how to figure out a chronology when clear dating clues are not present. Medievalists (of which I am one) have to do this a lot.
32/n
32/n
Well, it turns out that evolutionary biologists care a lot about change-over-time, too. Because that, in essence, is what evolution is: change over time. So they, too, have invested a lot of time & energy in thinking about how to date otherwise undatable events.
33/n
33/n
They use "molecular clocks." That is the assumption that change happens at the molecular level (& that's what genomes are: assemblies of molecules) at a regular rate. Except, apparently it doesn't. This is a matter biologists are still debating. Meanwhile, ...
34/n
34/n
... us historians need to continue to go about our business. So what I've been doing (while the biologists keep investigating their chronological measures) is to treat the SNP-derived dating estimates that biologists have proposed as rough estimates. And then I ask: So ...?
35/n
35/n
Which brings up back to the SNP Bingo card I showed before. Here it is again. I've written several times about the major polytomy (or Big Bang) that occurred in #YersiniaPestis' evolutionary history in the later Middle Ages. This was 1st postulated in 2012 by Cui et al.
36/n
36/n

There's a lot that could be said about the Big Bang (& in fact, I already have said a lot: academic.oup.com/ahr/article/12…). But the key thing to understand now is that it must have occurred before the well-documented BD that struck the Middle East & Europe starting in 1346.
37/n
37/n
Here's the dating question: How much earlier than 1346? There's also a geography question: Where did the Big Bang occur? Well, I've written about that, too, & postulated that it occurred (surprise!) in Kyrgyzstan, in/near the Tian Shan mountain range. Here's my 2020 map.
38/n
38/n

The present study by Spyrou & colleagues retrieves Y. pestis aDNA from 3 individuals from a gravesite at Kara-Djigach in northern Kyrgyzstan. These, of course, are dated burials. And the genome they have in their bodies seems to be pretty darn close to the genome predicted.
39/n
39/n
So, a "win" all around: the vague "pestilence" reported on the gravestones is confirmed as plague; the strain of Y. pestis in the bodies is close to the genomic signature as anticipated. And since the bodies are dated, we now know both place & time of the Big Bang. Right?
40/n
40/n
Well ....? This is where we get to that missing SNP.
41/n
41/n
It's important to recognize the different ways in which things can be missing. The 1st way--and this is a problem that all medievalists understand--is that what was once present in the past (a book, a church, the lyrics to a song) ..
42/n
42/n
... has been lost due to the passage of time. This seems to be the case with the quality of genetic data retrieved from the Kara-Djigach burials. Palaeogeneticists have more or less decided that, to be judged "good enough," the data they retrieve needs 3-fold coverage.
43/n
43/n
What's 3-fold coverage? The genome of #YersiniaPestis is ca. 4.6 million base-pairs long. A "complete" genome will have the bases in place for every point along the genome. aDNA has to work w/ degraded fragments. There will be some points of the genome w/ no coverage at all.
44/n
44/n
But if preservation is good, there may be positions along the genome where multiple fragments overlap to produce coverage at 5-fold, 10-fold. Think how excited you'd be to get up to 27-fold! Here's the reported coverage for the Kara-Djigach genomes.
45/n
45/n

What that chart indicates, in the "mean coverage" column, is that when all the reads from the several passes of both the BSK001 and BSK003 genomes were pooled together (highlighted in red), the best possible coverage for the genome as a whole was an average of 9.5-fold.
46/n
46/n
Now let's look at some SNPs in detail. Our earlier "SNP Bingo" card told us that we should look for three specific SNPs that defined Y. pestis' evolutionary history betw the divergence point of the pre-polytomy lineage & its "cousin", the still-extant lineage 0.ANT3.
47/n
47/n
These three SNPs are as follows:
- A1102174G
- T1251046C
- G2812384T
(The attached screenshot gives a quick explanation of how to "read" SNP labels.)
48/n
- A1102174G
- T1251046C
- G2812384T
(The attached screenshot gives a quick explanation of how to "read" SNP labels.)
48/n

Let's look at the data for the Kara-Djigach genomes & see what kind of quality they give us. This data is from Spyrou et al Supplementary Table 18: Phylogenetically diagnostic SNPs in ancient & modern Y. pestis genomes - detail of SNPs defining 0.ANT3 & pre-polytomy lineage.
49/n
49/n

And here's a closer look. Of the three SNPs that define the pre-polytomy lineage--the three SNPs on which the entire historical argument of this paper rests--only 1 SNP meets the 3-fold criterion in both the two Kara-Djigach genomes. The other two only have 1-fold coverage.
50/n
50/n

1-fold coverage is better than nothing, of course, so let's take this as a "win." After all, we're medievalists & know that oftentimes you have to make due w/ poor quality data. But, before you move away, glance at some of those other columns in the previous image.
51/n
51/n
I'll repeat it. Look at column G, LAI009, the now-famous Laishevo genome which has been said to be the earliest form of Branch 1 to arrive in Europe: the real "mother" of the BD as it would be experienced in western Eurasia. 3-fold coverage (at least) in every position.
52/n
52/n

Look, too, at "London-ES" and "Barcelona3031." Those are the genomes extracted, on the one hand, from the famous Black Death Cemetery in London (the 1st great success of the present research team) & from the basilica at Barcelona, where bodies were dumped into the walls.
53/n
53/n
So, where's the "missing SNP" we were supposed to be looking for? It's not even reported in this Table. As always with prestidigitation, you want to show onlookers the rabbit before you make it disappear. So they show us.
54/n
54/n

And then it's gone: "This SNP was found in a region with persistent multi-allelic sites; therefore, it is considered artefactual." The only other occasion I found SNP C3017633T mentioned was in this Supplementary figure 8.
55/n
55/n
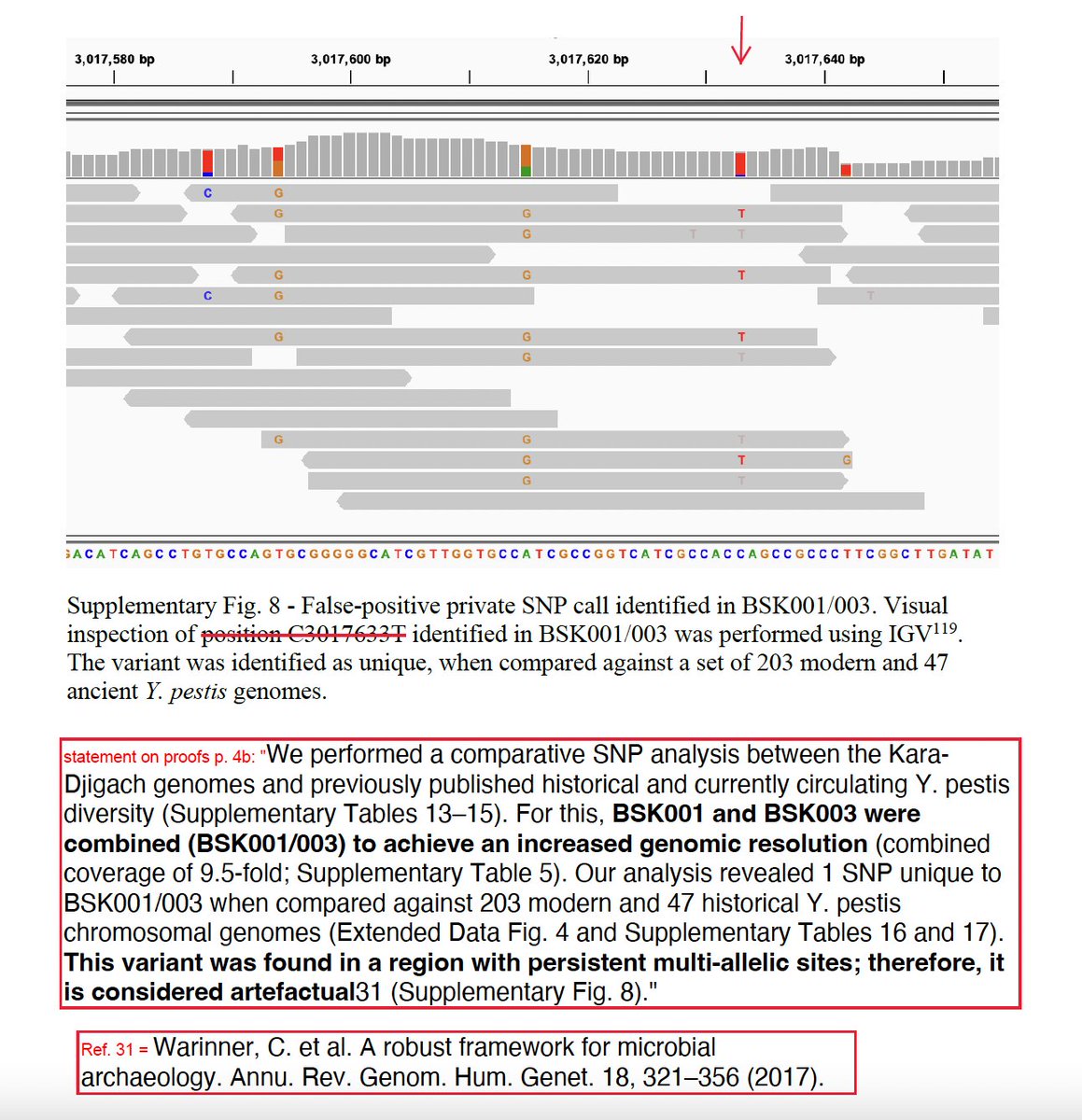
Okay, I'm a medievalist. I work w/ texts. Sometimes you find some data that seems tantalizing at first, but you end up having to reject it b/c its just too anomalous. But stop & think. We're talking about the birth of a new lineage (the pre-polytomy lineage), which is ...
56/n
56/n
... defined by 3 SNPs. Remember, that's just 3 base-pair positions on a genome that's 4.6 million basepairs long. And you've found a new SNP. AND YOU THROW IT OUT? Is it a poor quality SNP, like the other ones we looked at? No. It's got 9-fold coverage.
57/n
57/n
Here is what the historian expects. You don't throw out data w/o a very good reason. You have to explain that reason. Maybe calling something "artefactual" is good enough for biologists. It's not good enough for historians. And that's b/c too much hangs in the balance.
58/n
58/n
End Part III. On to Part IV: The Case of the Missing Genomes.
Let's say that "disappeared SNP", C3017633T, had been left in the assessment of these retrieved genomes from Kara-Djigach. What difference would that have made?
59/n
Let's say that "disappeared SNP", C3017633T, had been left in the assessment of these retrieved genomes from Kara-Djigach. What difference would that have made?
59/n
Well, since SNPs are used as rough dating measures for estimating time in #YersiniaPestis evolution, it would lengthen the overall period that it took for the Kara-Djigach strain (w/ 4 new SNPs)--as distinct from the pre-polytomy lineage (w/ 3 distinctive SNPs) to emerge.
60/n
60/n
[As an aside, I'd also point out that it would constitute a "Fifth Black Death"--a 5th lineage of plague that emerged out of whatever event caused the other 4 lineages to emerge after a sudden dispersal. There may, indeed, have been many more such dispersals.]
W/ or w/o a 4th SNP, we still have molecular clock problems. Does Y. pestis have a clock? I'd love to know, & I await better answers from the biologists. In the meantime, I would note this: marmots (plague's long-term hosts in the Tian Shan) seem to induce a much slower ...
61/n
61/n
... rate of change than other hosts. Look, for example, at how little 0.ANT3--the "cousin" of our pre-polytomy lineage--has evolved since it diverged from the former. In, say, 700 years, it has acquired only 15 unique SNPs. This phylogenetic tree (based on more samples ...
62/n
62/n
... of the strain than were used in the present study) has all its documented diversity crammed into a narrow window between 1928 & 2001. Again, this is the cousin of the lineage that led up to the polytomy. It's our closest model for how marmot strains behave.
63/n
63/n

With this (apparent) slow rate in mind, we can turn to the Case of the Missing Genomes. What genomes were these? Again, this disappearing act was done completely in public view. The study is based on 250 genomes: 203 modern ones + 47 retrieved from aDNA. Good.
64/n
64/n
But then look what happens when it comes time to do the dating analysis in fig. 3? These diagrams are based on only 167 genomes. Wait? What happened to the other 83?
65/n
65/n

I don't work w/ such statistical tools as BEAST & it may be there's some obvious explanation for the culling of a full third of the samples used elsewhere for analysis. But I do know what it looks like: all the samples older than cousin 0.ANT3 were left out of the analysis
66/n
66/n
What it looks like is an effort to push the results toward a more modern "birth date" of both the pre-polytomy lineage & the Big Bang itself. The analogy I came up w/ was this.
67/n
67/n
Say I'm going to do a study about the emergence of X phenomenon in the 20th century. I collect relevant samples from all 100 years of the century. But then, when it came time to assess the actual date of emergence, I left out all the samples from before 1950.
68/n
68/n
As with the case of the disappeared SNP, such unexplained analytical shifts are anathema to historical method.
69/n
69/n
Which brings me to my final point: the main argument of the study is not only to claim they have established the presence of plague in this 14thC burial ground in Kyrgyzstan, & that they have established its (near) identity to the pre-polytomy strain that was predicted, ...
70/n
70/n
... but also that they have *proven* the date of Big Bang itself, placing it firmly in the 14th century. Why should such a strong argument be made to hang on (as we've seen) less than robust genetic data? Why, too, is such a strong argument being put forward ...
71/n
71/n
... when the latest work by historians, who have been paying close attention to all these developments in genetics, pushes the main period of plague's long-distance movements into the 13th century, not the 14th?
72/n
72/n
As I said at the outset, I am addressing this thread not simply to my fellow historians, but to the editors at @Nature. I may be missing something obvious, but, having read the peer review reports, I know that your four readers missed the points I have made here.
73/n
73/n
The paper by Spyrou et al ends w/ welcome words on the need for interdisciplinarity. But that cannot be effectuated solely by having single individuals as "tokens" on research teams. That needs to be effectuated at the review stage as well.
74/n
74/n
This study has disappeared not simply 1 SNP & 87 genomes, but a whole century of history! If palaeogenetics is going to claim to be a historicist science, then it needs to make its results intelligible to its real audience: fellow historians.
75/end
75/end
This thread has 75 tweets; if they don't all display, hit "Show replies."
Corrigenda to the above thread:
tweet #15: "diseased", should be "deceased"
tweet #23: should be "that it was plague"
tweet #75: "87 genomes", should be "83 genomes" (See tweets #64 & 65 above)
Corrigenda to the above thread:
tweet #15: "diseased", should be "deceased"
tweet #23: should be "that it was plague"
tweet #75: "87 genomes", should be "83 genomes" (See tweets #64 & 65 above)
• • •
Missing some Tweet in this thread? You can try to
force a refresh




