
🔴 A lot of discussions have come up on the BA.2.75 variant of #SARSCOV2. A continuously updated 🧵on emerging evidence on this variant. 

This variant was highlighted for designation recently. Majority of genomes have come from India 🇮🇳, apart from many other countries. github.com/cov-lineages/p…
Why is this lineage interesting?
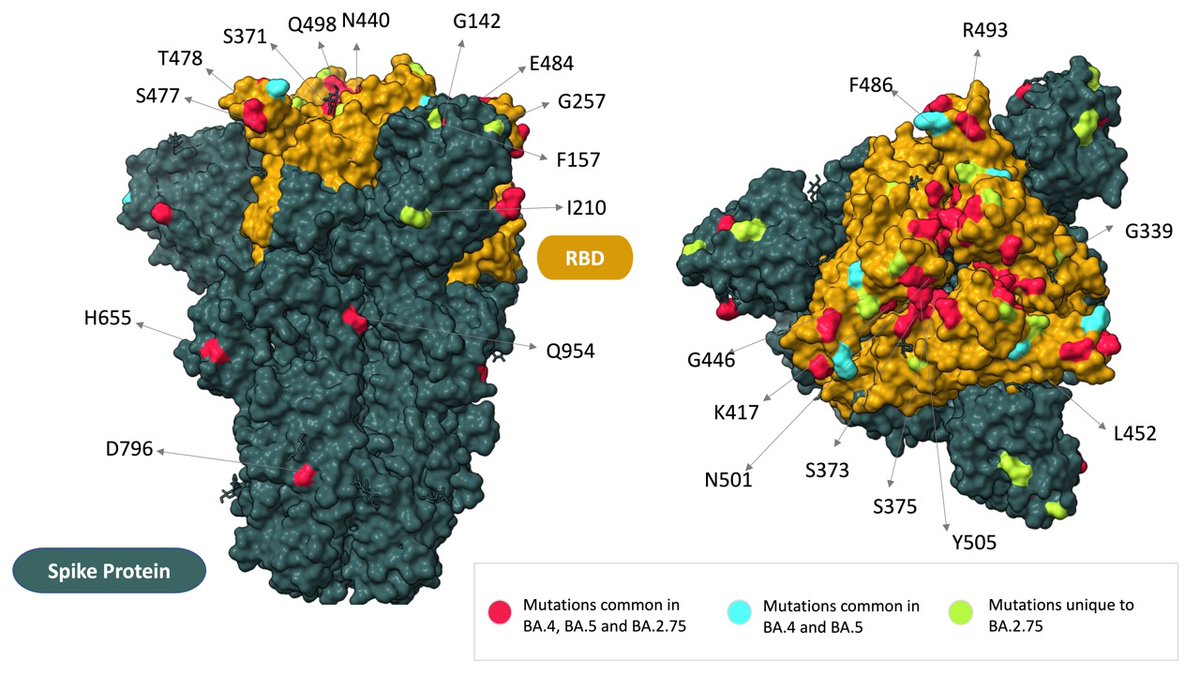
⚪ Interesting because it has 9 unique changes in the spike protein incl. a reversion compared to the BA.4 and BA.5 subvariants of omicron now spreading widely across the globe.
Schema @mercy_rophina
⚪ Interesting because it has 9 unique changes in the spike protein incl. a reversion compared to the BA.4 and BA.5 subvariants of omicron now spreading widely across the globe.
Schema @mercy_rophina

If you map the mutations to the spike protein, this becomes a bit clear. G446S is in the RBD which binds to the human receptor and also associated with major immune (Ab) escape
Plot @mercy_rophina
Plot @mercy_rophina
🔴 Immune Escape
G446S can possibly influence both immune escape and ACE2 binding. Given data at hand, it is very likely that while this might decrease the binding efficiency, could contribute to significant immune(ab) escape based on data from @jbloom_lab
jbloomlab.github.io/SARS-CoV-2-RBD…
G446S can possibly influence both immune escape and ACE2 binding. Given data at hand, it is very likely that while this might decrease the binding efficiency, could contribute to significant immune(ab) escape based on data from @jbloom_lab
jbloomlab.github.io/SARS-CoV-2-RBD…
This could mean Reinfections vaccine breakthrough infections could drive the spread of BA.2.75.
@jbloom_lab has an extensive thread on this 👇
@jbloom_lab has an extensive thread on this 👇
https://twitter.com/jbloom_lab/status/1542526095299203074?t=JxGTo1lSNu6EkvYSdVLwKg&s=19
⚪ Growth advantage
While the number of genomes from India in public domain for the variant is quite small to preclude assessments, the rapid increase in recent weeks (albeit small numbers in very small number of genomes) suggest it might have a growth advantage ...
While the number of genomes from India in public domain for the variant is quite small to preclude assessments, the rapid increase in recent weeks (albeit small numbers in very small number of genomes) suggest it might have a growth advantage ...
compared to BA.2 and sublineages.
⚪ Point to note here is BA.2 and sublineages are the ones prevalent in India presently and BA.4 and BA.5 has not caught up significantly in Maharashtra where most of the BA.2.75 has come up. So essentially it is competing with other BA.2.x now.
⚪ Point to note here is BA.2 and sublineages are the ones prevalent in India presently and BA.4 and BA.5 has not caught up significantly in Maharashtra where most of the BA.2.75 has come up. So essentially it is competing with other BA.2.x now.
🔴 Is there a need to panic now ?
NO, as the variant is NOT YET suggested to cause severe disease or increased mortality.
Point to keep in mind is that the variant is continuously evolving and accumulating more mutations and it is too early to jump into conclusions
NO, as the variant is NOT YET suggested to cause severe disease or increased mortality.
Point to keep in mind is that the variant is continuously evolving and accumulating more mutations and it is too early to jump into conclusions
⚪ a much broader & stronger genomic surveillance, epidemiology and more importantly, availability of data for analysis is key to understand the picture in a better way.
🔴 How can you protect yourself and your near and dear ?
😷 better masks , more masks
☁️ better ventilation
💉 complete the vaccine schedule for all.
😷 better masks , more masks
☁️ better ventilation
💉 complete the vaccine schedule for all.
• • •
Missing some Tweet in this thread? You can try to
force a refresh








