
Our many-years-in-the-making EpiC Dog epigenome paper is finally out as preprint @biorxivpreprint
Close collaboration with our lab's lead bioinformatician @new__hong & MBD-seq aficionado @MethylAReum, spearheaded by our versatile PI @je_yoel @SNUnow #Epigenome #Genomics #Dog A🧵
Close collaboration with our lab's lead bioinformatician @new__hong & MBD-seq aficionado @MethylAReum, spearheaded by our versatile PI @je_yoel @SNUnow #Epigenome #Genomics #Dog A🧵
https://twitter.com/biorxivpreprint/status/1550715423003496448

While still in early stage format, EpiC Dog is a culmination of our efforts to annotate functional elements of dog genome enabled by chromatin states discovery and profiling super-enhancer and DNA methylome landscapes across multiple tissue types. See also BarkBase and DoGA /2 

Modern dogs are champions of modern genetics. They empower us with their evolution and diversification of phenotypic traits from morphology to behavior. One reason dog genomics has become >popular compared to genomic studies of other domesticated mammals, but who's comparing? /3
On-going efforts to catalog dog epigenome currently lack integrative genome-wide datasets of chromatin states, super-enhancers, and DNA methylomes. So we bring to the table via EpiC Dog integrated datasets of RNA-seq, 5 histone marks ChIP-seq, MBD-seq, and ATAC-seq /4
Aside from integration of these multi-type datasets, we performed comparative epigenome analyses utilizing ENCODE datasets for human and mouse. It's still surprising to us that we find more epigenomic similarities bet human and dog than bet human and mouse /5
Comprehensive transcriptomics of 11 tissue types. We deconvoluted transcript abundance (>22,000 protein-coding genes & >45,000 transcripts) in dog by performing multiple clustering strategies and defined species and tissue specificities. We also GO-ed all of them for function /6 

And then the highlight of the study: genome-wide chromatin state discovery! We described a 13-state model informed by 5 histone H3 marks using ChromHMM. This model consists of 8 active states, 4 repressive states, and a quiescent state. /7 
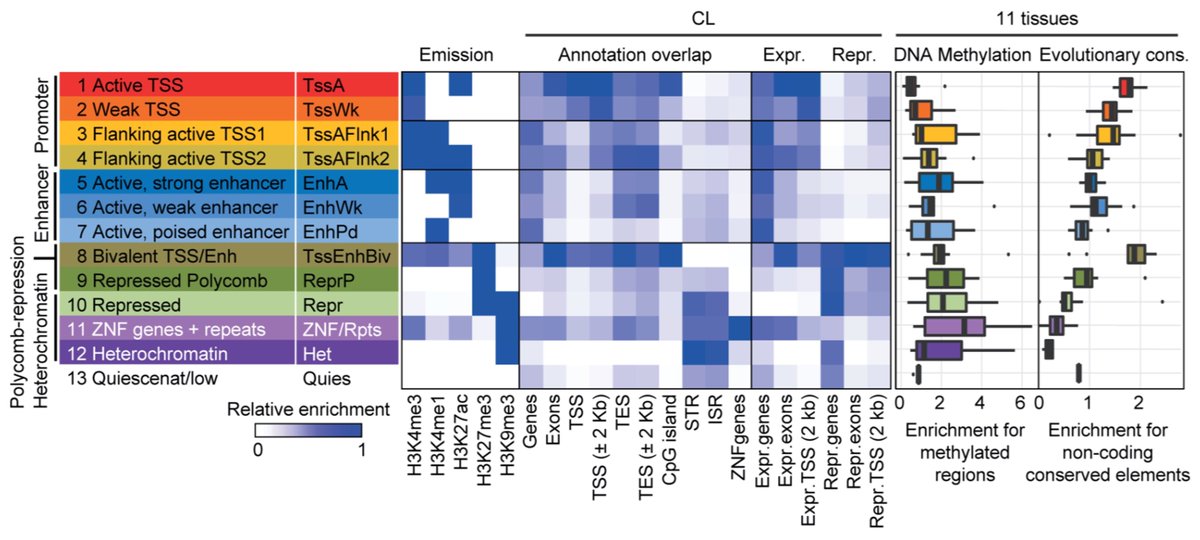
We expanded the dog epigenome profile by performing cross-species analyses of conservation by synteny and matched inter-tissue variation. These data point to a higher chromatin landscape similarity bet human and dog than human and mouse. /8 

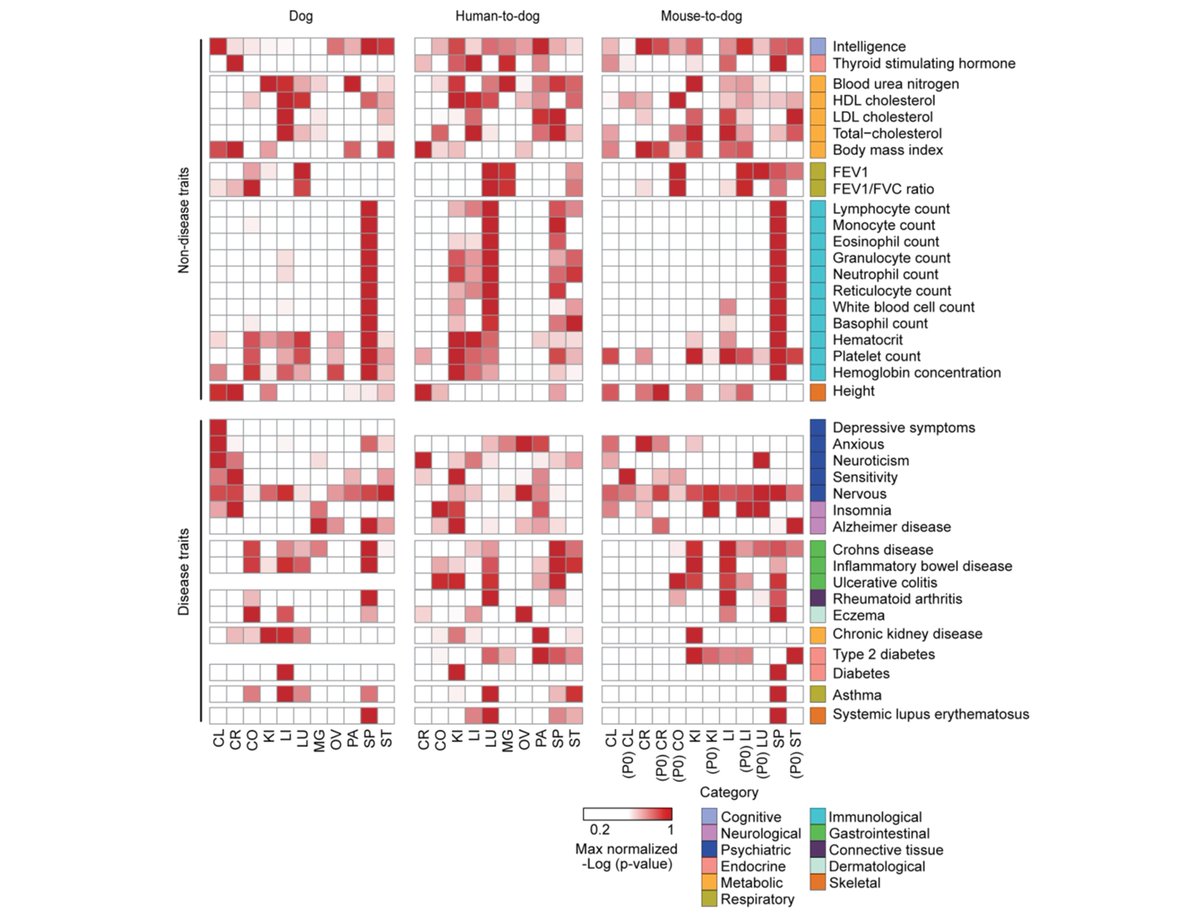
We next studied regulatory annotation enrichments of phenotype-associated variants from GWAS summary statistics for 49 phenotypes. We retrieved a total of 687 significant associations between 24 tissues and 38 complex phenotypes in dog, human-dog, and mouse-dog alignments. /9 
We next probed tissue identity and function by mapping super-enhancer (domain) landscapes and gene linking. Aside from confirming tissue identities, we mainly focused on SE domains and found 6654 regions with distinct mode-of-actions other than tissue specificity. /10 

Finally, we profiled global DNA methylome landscapes and delineated inter-tissue signatures. We annotated common methylated CMRs and tissue-specific methylated tsDMRs. We identified >7.1mil CMRs and >20.4k tsDMRs and performed gene mapping on these regions to annotate DMGs. /11 

To wrap up, we made an open-access repository and a genome browser to allow for mapping and visualization of our integrated epigenome data. These efforts are preliminary and we are expanding to make a data portal and add new datasets. /12 

That's it! I took a longer-than-expected break from cancer research to work on this project. Thanks to our collaborators and other people who helped along the way. Acknowledging our NRF grants and our recent SRC funding w/c transformed our lab into becoming CDRC /13 

And oh! To give faces to the co-first-authors and our PI 🧑🔬👩🔬🔬🧬, here's an impromptu photo taken on our lab balcony (forget my hair pls😥) @new__hong @MethylAReum @je_yoel Also want to acknowledge co-authors @ATACseek twitterless Kang-Hoon Lee, Kyung-Ju Shin & Keunsoo Kang. /14 

• • •
Missing some Tweet in this thread? You can try to
force a refresh



