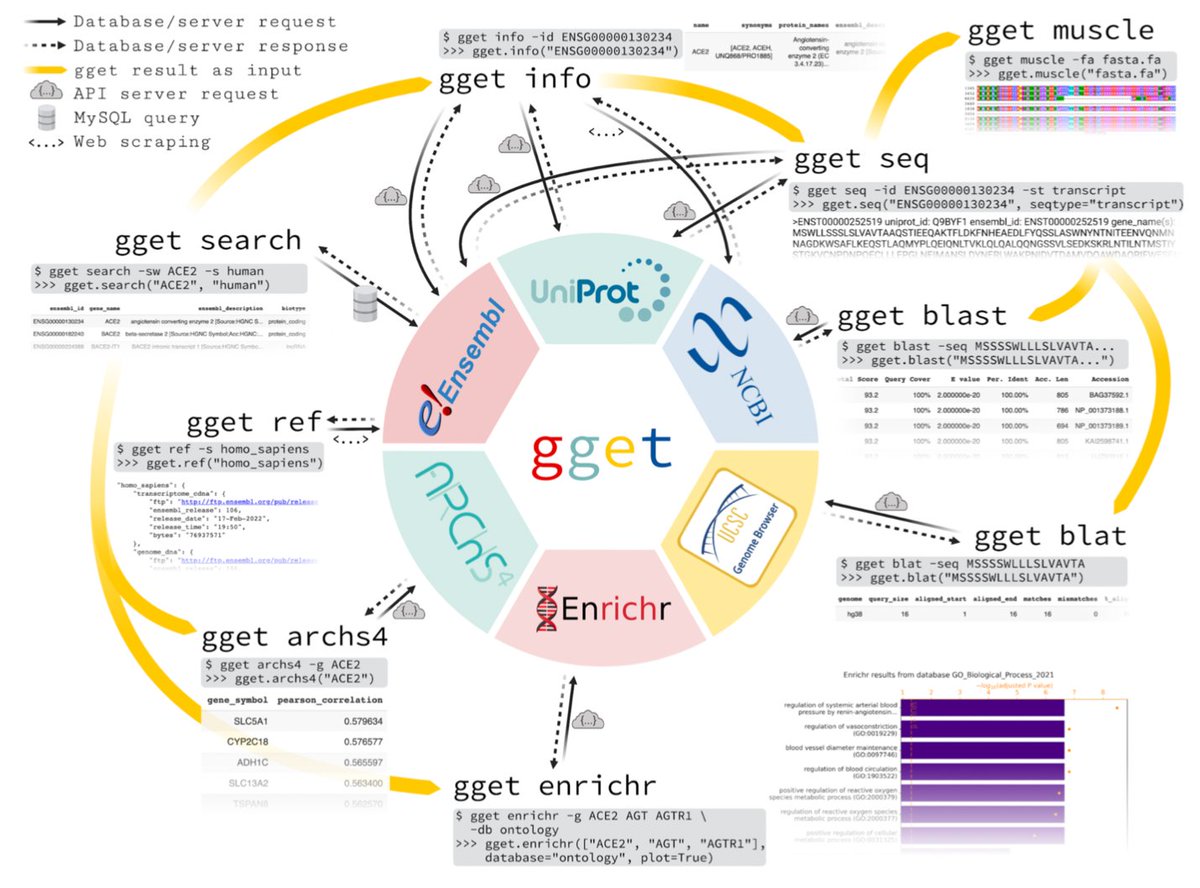
Tons of exciting new single-cell genomics tools have been showcased at #bioc2022 this week. Today @LambdaMoses presented SpatialFeatureExperiment, an S4 class extending SpatialExperiment, facilitating geospatial stats for spatial #scRNAseq using Voyager github.com/pachterlab/Voy… 1/ 

The design of SpatialFeatureExperiment and the plans for Voyager were formed from a careful study that @LambdaMoses conducted of the spatial transcriptomics field (published as the "Museum of Spatial Transcriptomics"): nature.com/articles/s4159… 2/
While there are several analysis tools for spatial transcriptomics data, and extensions of #scRNAseq platforms such as Seurat for spatial data, they have limitations in terms of the methods they implement from the field of geospatial statistics. 3/
Also there has also been a breakdown in #RStats when it comes to single-cell genomics, limiting the utility of SingleCellExperiment, and SpatialExperiment which extends it. 4/
https://twitter.com/rmassonix/status/1550107819738906624?s=20&t=VLXTPXnUekq69ThpVknN0Q
By developing Voyager on SpatialFeatureExperiment, which extends SpatialExperiment, hopefully single-cell genomics in #Rstats can re-center itself on interoperable classes. Voyager is just being launched by @LambdaMoses, and will mature as spatial genomics technologies mature. 5/
The plan is not only to have a framework applicable to the many different assays that have been / are being developed, but also to incorporate the large body of work in geospatial statistics that has not been fully explored in the single-cell genomics community. 6/6
• • •
Missing some Tweet in this thread? You can try to
force a refresh