
[1/5] How can we improve pathologist efficiency in screening of normal colon biopsies? Take a look at our new #preprint that tackles this issue using #interpretable #AI 🔬 🚀 @PathLAKE_CoE
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
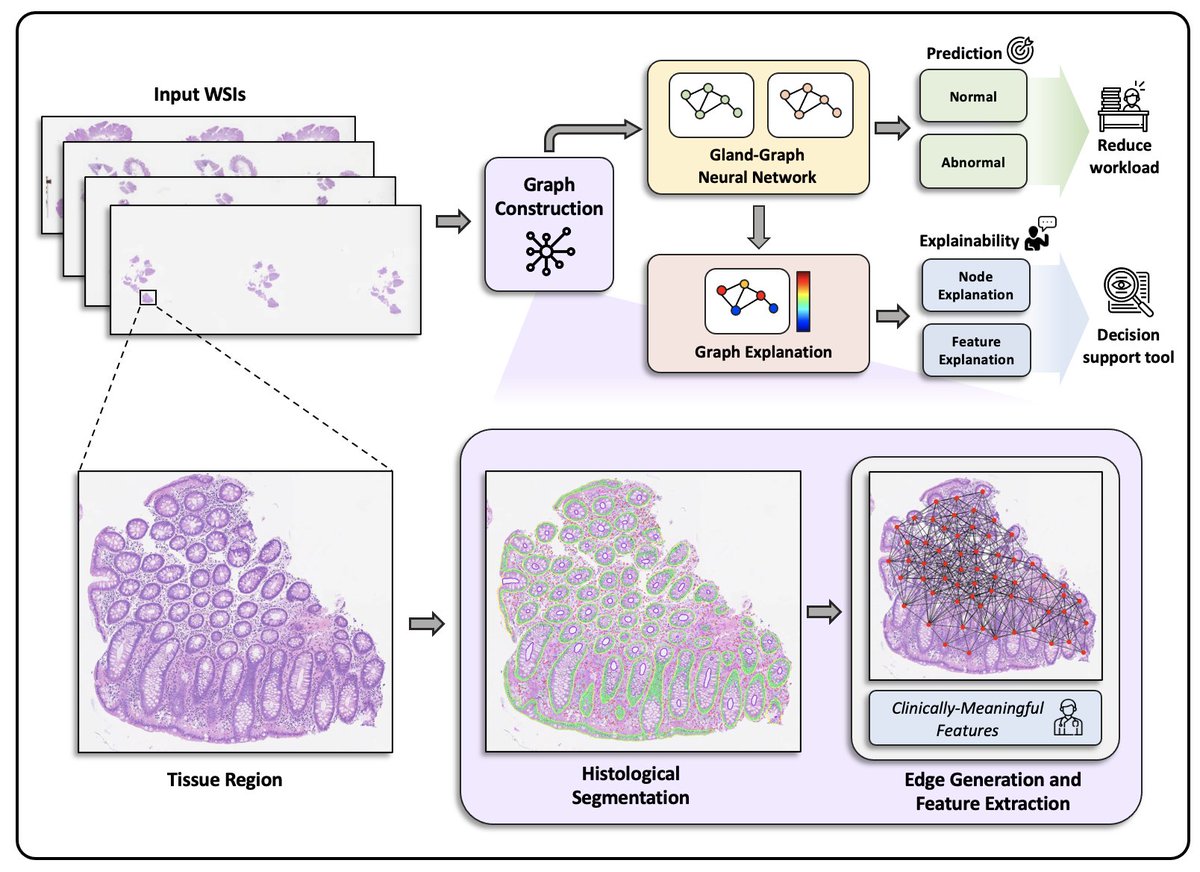
@PathLAKE_CoE @jnkath @AI4Pathology @NormanZerbe @anantm @mitkoveta @olkazuraw @GuillaumeJaume @pushpak_pati @jana_lipkova @nadeem_lab [2/5] Our approach, named #IGUANA, uses #pathologist-derived #interpretable features within a geometric learning framework to categorise biopsies as normal or abnormal (containing both non-neoplastic and neoplastic conditions).
[3/5] IGUANA achieves 0.98 AUC-ROC on an internal cohort and 0.97 AUC-ROC on three external cohorts. Our analysis shows that at a high sensitivity threshold of 99%, the proposed model can, on average, reduce the number of normal slides to be reviewed by a pathologist by 55%.
[4/5] A key advantage of #IGUANA 🦎 is its ability to provide an #explainable output, highlighting potential abnormalities in a #WSI as a heatmap overlay, in addition to numerical values associating the model prediction with various histological features. 

[5/5] Check out our interactive demo (not optimised for smartphone) online at iguana.dcs.warwick.ac.uk 💻
• • •
Missing some Tweet in this thread? You can try to
force a refresh





