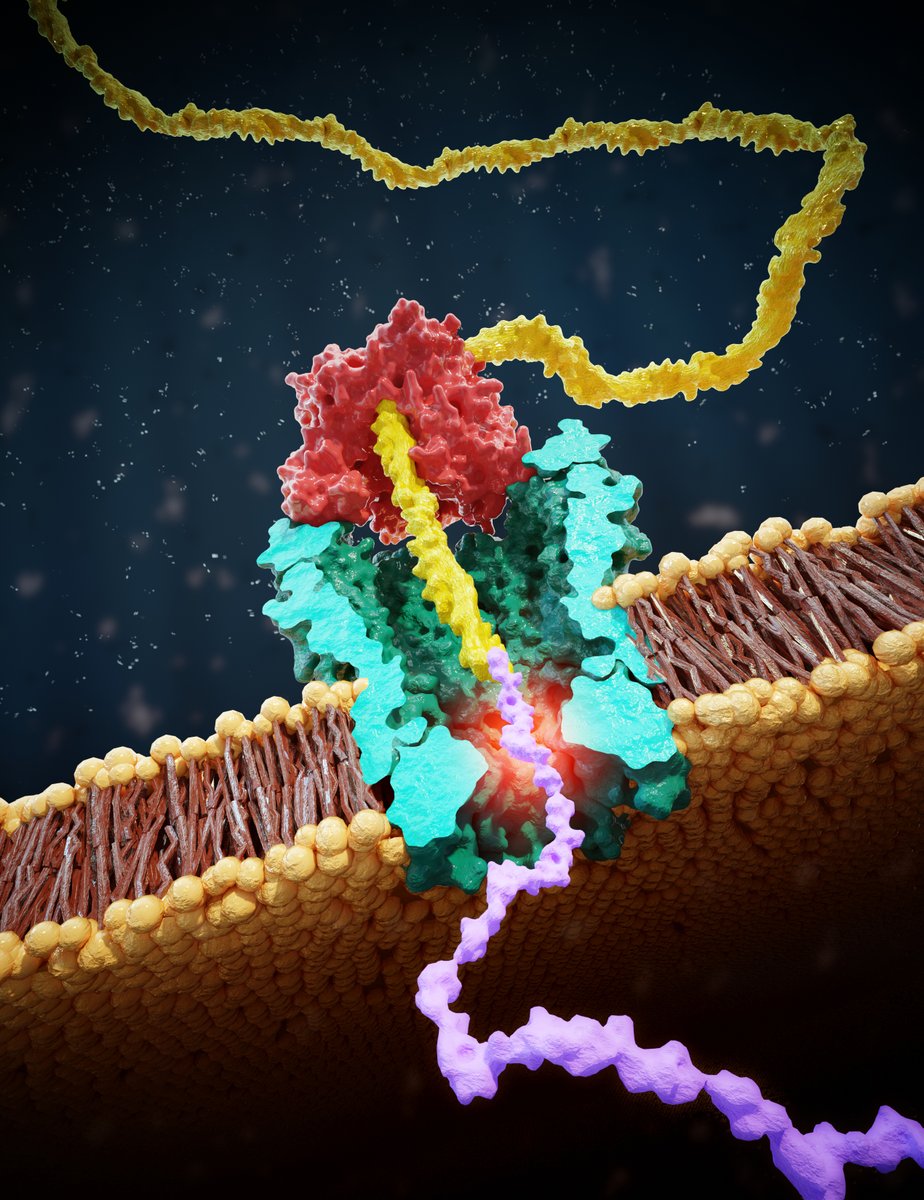
Protein sequencing is a big deal and goes way beyond DNA sequencing. While we have ~20000 protein-coding genes, we have _millions_ of protein variants, mainly because of post-translational modifications that attach a side group to amino acids.
1/
1/

Phosphorylation is the most frequent PTM, and of particular interest, as dysregulation of phosphorylation pathways is linked to many diseases including cancers, Parkinson’s, Alzheimer’s, and heart disease.
2/
2/

We lack techniques to measure such PTMs on the single-molecule level! 😳
=> We need new single-molecule techniques
(Mass spectrometry requires typically more than a billion copies and often struggles to identify the correct position of a PTM between multiple candidate sites)
3/
=> We need new single-molecule techniques
(Mass spectrometry requires typically more than a billion copies and often struggles to identify the correct position of a PTM between multiple candidate sites)
3/

We now put a new #CDlab paper on @biorxivpreprint where we use our recently introduced nanopore single-protein scanning method (see here
4/
https://twitter.com/cees_dekker/status/1456323490647212033?s=20&t=5Ci_drI-nwYZvTKErh70Mw) to detect single PTMs within single molecules: biorxiv.org/content/10.110…
4/

We found that this approach allows extremely sensitive measurements that can clearly distinguish peptides with or without a single small PTM, for example the presence of 1 phosphate group (only 5 atoms!) on the immunopeptide BCAR3, a promising neoantigen for immunotherapy.
5/
5/

The method also can sensitively detect and discriminate single phosphate groups within individual peptides, for example 2 PTMs that are only 3 amino acids apart on another clinically relevant immunopeptide, βCAT
6/
6/

The accuracy of detection these PTMs is excellent, about 95% accuracy, even for individual reads of single molecules (and this can be increased with re-reading)
7/
7/

Finally, we did Markov-chain Monte Carlo calculations to model the behavior of these mixed-charged peptides in nanopores, and we find that the charged phosphate PTMs ‘lag and hop’ from the pore moth into the pore lumen, which explains the current profiles
8/
8/

All this work was done by PhD student Justas Ritmejeris and postdocs Ian Nova and Henry Brinkerhoff who now is at the lab of Jens Gundlach.
Read all about it in our preprint:
biorxiv.org/content/10.110…
9/
Read all about it in our preprint:
biorxiv.org/content/10.110…
9/

• • •
Missing some Tweet in this thread? You can try to
force a refresh