
hoofd & hart | nanotech & biofysica | TUDelft & prive | geloof & wetenschap | boeken & muziek | schepping & evolutie | Jezus = Heer
How to get URL link on X (Twitter) App

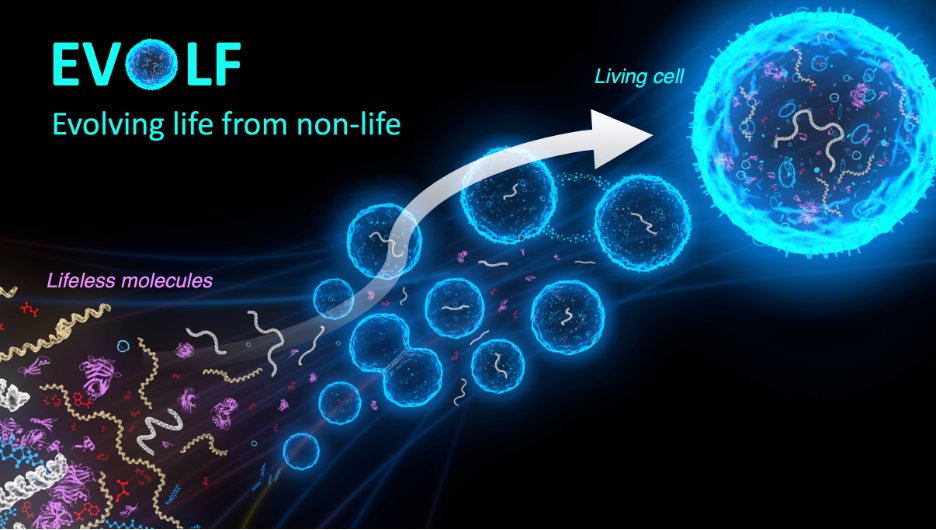
 This project aims to address some of the biggest questions: What is life? How do living systems differ from non-living ones? Can we create living cells from lifeless molecules? EVOLF will take an experimental approach and build synthetic cells from the bottom up, from molecules.
This project aims to address some of the biggest questions: What is life? How do living systems differ from non-living ones? Can we create living cells from lifeless molecules? EVOLF will take an experimental approach and build synthetic cells from the bottom up, from molecules. 
 But here's a new twist!
But here's a new twist!

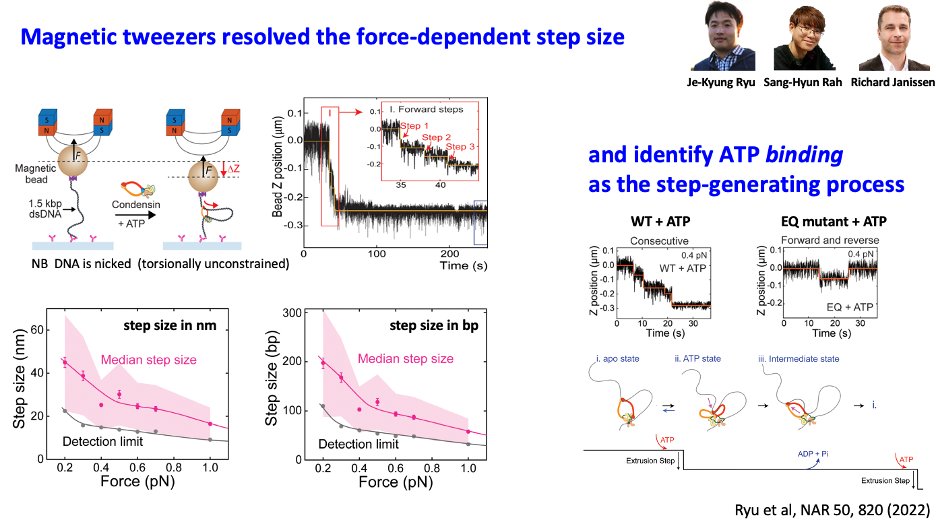
 Background of this paper is interesting as the 4 authors met at a Titisee conference. Given that each of us had a favorite model (scrunching, swing&clamp, hold&feed), we disagreed.
Background of this paper is interesting as the 4 authors met at a Titisee conference. Given that each of us had a favorite model (scrunching, swing&clamp, hold&feed), we disagreed.

https://twitter.com/cees_dekker/status/1630580415483265024As a follow up:



 Phosphorylation is the most frequent PTM, and of particular interest, as dysregulation of phosphorylation pathways is linked to many diseases including cancers, Parkinson’s, Alzheimer’s, and heart disease.
Phosphorylation is the most frequent PTM, and of particular interest, as dysregulation of phosphorylation pathways is linked to many diseases including cancers, Parkinson’s, Alzheimer’s, and heart disease.


 @arxiv Such rotors are inspired by the awesome F0F1 ATPase motor protein in our cells. Here, a proton gradient drives rotation of F0 which induces conformational changes in F1 that catalyze production of ATP, which is the fuel for most processes in our body.
@arxiv Such rotors are inspired by the awesome F0F1 ATPase motor protein in our cells. Here, a proton gradient drives rotation of F0 which induces conformational changes in F1 that catalyze production of ATP, which is the fuel for most processes in our body.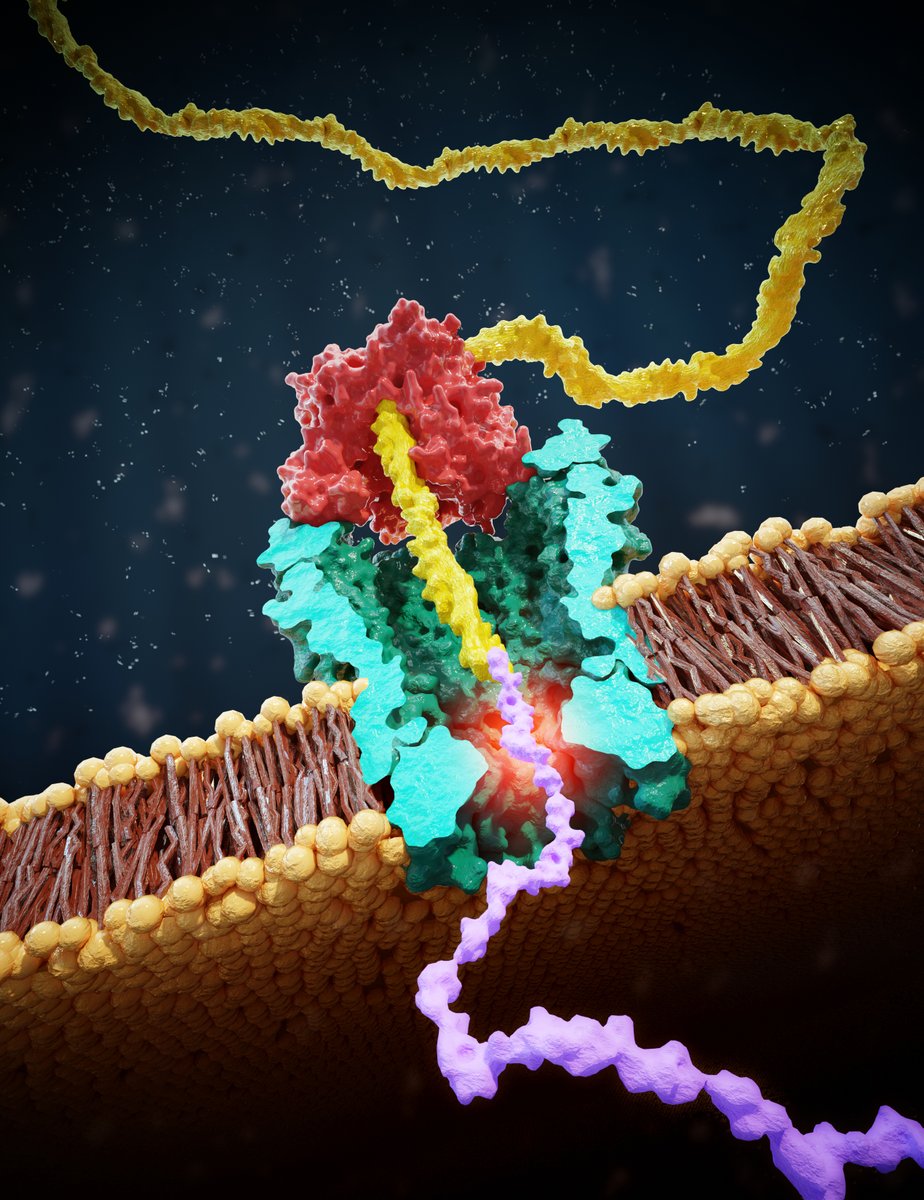
 @ScienceMagazine @aksimentievLab Here’s the link to this paper in @ScienceMagazine entitled “Multiple re-reads of single proteins at single-amino-acid resolution using nanopores”: science.org/doi/10.1126/sc…
@ScienceMagazine @aksimentievLab Here’s the link to this paper in @ScienceMagazine entitled “Multiple re-reads of single proteins at single-amino-acid resolution using nanopores”: science.org/doi/10.1126/sc… 