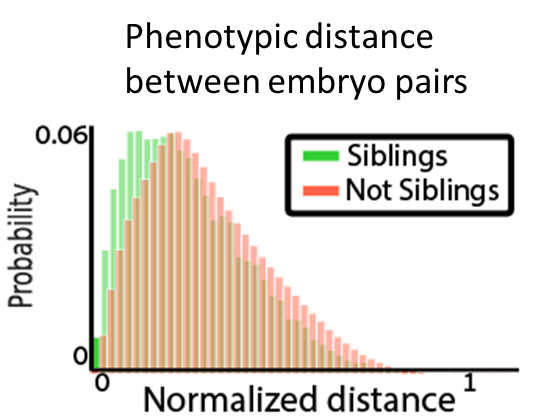
Very happy to share our new @biorxivpreprint showing that phenotypic correlations between in vitro fertilization (#IVF) sibling embryos provide predictive value regarding an embryo’s future implantation potential!
biorxiv.org/content/10.110…
🧵
1/n
biorxiv.org/content/10.110…
🧵
1/n

The implantation potential of transferred embryos was associated with the phenotype of their non-transferred “sibling”. Siblings’ phenotypes contribute to image-based machine learning embryo implantation prediction for models trained with morphology/morphokinetic features!
3/n
3/n

The fraction of sibling embryos reaching blastulation came up as the most important feature for prediction of implantation outcome in a model trained with morphology, morphokinetics, oocyte age & cohort features! 5/10 top features were attributed to the cohort’s siblings!
4/n
4/n

Embryos that were “rescued” by the cohort features, meaning correctly classified only by the classifier that had access to cohort information, relied on the two top ranked cohort features, and *only* “rescued” implanted embryos erroneously classified as non-implanted 🤔
5/n
5/n

We think that the model was able to correct negative-to-positive but not positive-to-negative predictions because failed implantations are caused either by defected embryos or maternal clinical properties translating to ambiguous labels that mislead the classifier
6/n
6/n

Altogether, we suggest that the #ML uncertainty in the transferred embryo is reduced by noise reduction from the multiple correlated (sibling) instances.
Or in other words, @Giannis_An34 could be predicted to be a basketball player according to his siblings 😉
7/n
Or in other words, @Giannis_An34 could be predicted to be a basketball player according to his siblings 😉
7/n

This is an example of semi-supervised learning: a small fraction of labeled (transferred embryos) and vast unlabeled (non-transferred siblings) observations. Unlike traditional semi-supervised learning, here the unlabeled and labeled observations are associated.
8/n
8/n

In summary, inclusion of cohort features contribute to machine learning models trained with embryo-derived features. Since the siblings’ data are routinely collected, incorporating them in #AI-driven embryo implantation prediction can have direct implications in the clinic!
9/n
9/n

Project led by (now graduate M.Sc.) Noam Tzukerman with @oded_rotem and our collaborators @DaniellaGilboa and team at @AiVFtech!
We are happy to hear thoughts and feedback :-)
10/n
We are happy to hear thoughts and feedback :-)
10/n

• • •
Missing some Tweet in this thread? You can try to
force a refresh