QuPath v0.4.0 is now available!
Download it at qupath.github.io
#opensource #bioimageanalysis #digitalpathology #java #javafx
Download it at qupath.github.io
#opensource #bioimageanalysis #digitalpathology #java #javafx

There are so many new things that it'll take some days to describe even just the main ones.
For now, I'll start by mentioning a few of the user interface improvements.
QuPath v0.4.0 is more welcoming than previous versions... and a bit more stylish.
For now, I'll start by mentioning a few of the user interface improvements.
QuPath v0.4.0 is more welcoming than previous versions... and a bit more stylish.
In fact, you can even style it in your own unique way if you really want to, thanks to #javafx & css (ideally in a nicer way than in my screenshots).
qupath.readthedocs.io/en/0.4/docs/re…

qupath.readthedocs.io/en/0.4/docs/re…


Confusing things have been (I hope) reduced.
Lots of options & dialogs have been revised & updated to be more intuitive & user-friendly.
Lots of options & dialogs have been revised & updated to be more intuitive & user-friendly.
Measurement tables now include images, annotations can have descriptions, and it's possible to undock the tabs on the left so you can see more things at once.
If multidimensional images are your thing, you should find annotating across z-stacks & timepoints to be easier - thanks to new commands to copy annotations between slices.
As before, you can use arrow keys to navigate between slices.
As before, you can use arrow keys to navigate between slices.
Transferring annotations between images is also much easier, since objects support copy & paste.
In fact, this uses text (GeoJSON) so you can even copy between software.
If you need to fix the alignment, interactively adjust one annotation & apply the transform to all of them
In fact, this uses text (GeoJSON) so you can even copy between software.
If you need to fix the alignment, interactively adjust one annotation & apply the transform to all of them
(I've run out of gifs & time, so I'll add more tomorrow)
Time for some more things in v0.4.0...
The script editor is a bit prettier and lot more helpful.
For starters, there’s a ‘Run’ button, 'File > Recent scripts...' menu, logged messages are in color, and a timer shows how long a script has been running.
The script editor is a bit prettier and lot more helpful.
For starters, there’s a ‘Run’ button, 'File > Recent scripts...' menu, logged messages are in color, and a timer shows how long a script has been running.
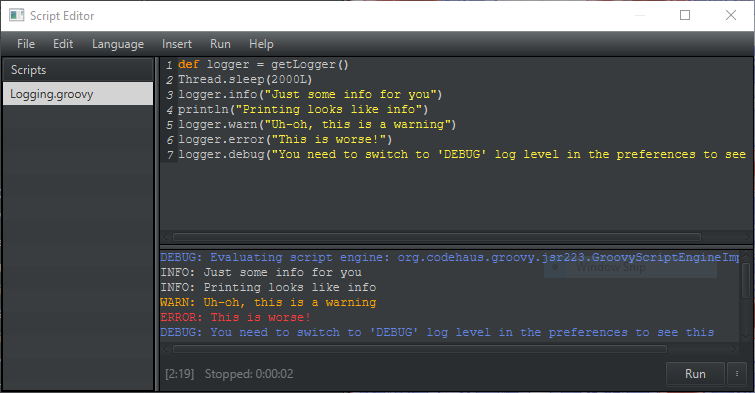
Press Ctrl/Cmd+Space to get completions for built-in (static) methods.
It’s not full autocomplete, but it helps with common things.
It’s not full autocomplete, but it helps with common things.
For less common things, or if you just want more info, the javadocs are now on hand.
These should describe every public method in QuPath that you might want to use in a script.
You can also find them at qupath.github.io/javadoc/docs/
These should describe every public method in QuPath that you might want to use in a script.
You can also find them at qupath.github.io/javadoc/docs/
There’s also basic syntax highlighting for more kinds of interesting files, not just Groovy scripts.
You can even work with JSON, YAML… even Markdown.
You can even work with JSON, YAML… even Markdown.
Bonus hack: I don’t *really* recommend it, but if you download Jython-standalone from jython.org/download you can drag it onto QuPath and start scripting with Jython instead of Groovy.
(Note this is limited to Python 2.x syntax & won’t give access to things like NumPy etc.)
(Note this is limited to Python 2.x syntax & won’t give access to things like NumPy etc.)

But scripting improvements aren’t limited to the editor.
A couple more changes should make Groovy scripting *much* more powerful, although it might take a little bit of time to prove that…
A couple more changes should make Groovy scripting *much* more powerful, although it might take a little bit of time to prove that…
One is that objects now have IDs.
This means that if you want to combine QuPath with R/Python/MATLAB/sth else (e.g. to do some fancy clustering), there’s a way to relate everything back to the original image - without needing to grapple with coordinates/centroids/other hacks.
This means that if you want to combine QuPath with R/Python/MATLAB/sth else (e.g. to do some fancy clustering), there’s a way to relate everything back to the original image - without needing to grapple with coordinates/centroids/other hacks.
The other is that objects have been upgraded to make accessing & updating measurements & classifications much more intuitive.
Combined with extra Groovy tricks, this can make powerful scripts much shorter & more readable – especially for complex/multiplex images.
Combined with extra Groovy tricks, this can make powerful scripts much shorter & more readable – especially for complex/multiplex images.
There are more details of the changes & thinking behind them at github.com/qupath/qupath/…
But if you don’t like scripting, the good news is that some things that used to require scripting now don’t.
For example, the new ‘Signed distance to annotations 2D’ command calculates distances to annotation boundaries, both from inside & out.
For example, the new ‘Signed distance to annotations 2D’ command calculates distances to annotation boundaries, both from inside & out.

Or you can make anything a Tissue Microarray with ‘TMA > Specify TMA grid’.
It’s no longer necessary to rely exclusively on the dearraying algorithm, or hack-y script.
It’s no longer necessary to rely exclusively on the dearraying algorithm, or hack-y script.
Beyond all that, there’s an experimental Apple silicon build for users of recent Macs.
There are a few caveats/limitations described at qupath.readthedocs.io/en/0.4/docs/in…
But if you’re ok with those, I’ve found things like cell detection run much faster than using the Intel build (~30%)
There are a few caveats/limitations described at qupath.readthedocs.io/en/0.4/docs/in…
But if you’re ok with those, I’ve found things like cell detection run much faster than using the Intel build (~30%)
And lastly for today, there’s now support for DICOM whole slide images.
That one is all thanks to everyone involved in updating the @bioformats plugin - QuPath just uses the latest version & benefits from that
That one is all thanks to everyone involved in updating the @bioformats plugin - QuPath just uses the latest version & benefits from that
https://twitter.com/bioformats/status/1468988615954939907?s=20&t=HZqmjSx6DwP6d_VRV41Wow
More to come tomorrow...
Got delayed, back now with more new things in v0.4.0...
QuPath has been used quite a lot by AI folks because it has some pretty nice annotation tools, as well as the ability to export in lots of customized ways.
QuPath has been used quite a lot by AI folks because it has some pretty nice annotation tools, as well as the ability to export in lots of customized ways.
https://twitter.com/petebankhead/status/1295965136646176768?s=20&t=zxPKZT2TuArSgGcuXPdzQw
But one big thing that was missing was the ability to run deep learning models from within QuPath.
It's been *technically* possible for quite a while (scripts make most things possible), but prohibitively awkward.
It's been *technically* possible for quite a while (scripts make most things possible), but prohibitively awkward.
https://twitter.com/petebankhead/status/1406898551439998980?s=20&t=f5GUwazOJgWn3OOQaztYng
v0.4.0 brings QuPath a lot closer to supporting deep learning routinely, with the help of @deepjavalibrary
It's still early - the focus has been on the unglamorous make-it-possible stuff - but I think it can already be pretty useful.
It's still early - the focus has been on the unglamorous make-it-possible stuff - but I think it can already be pretty useful.
https://twitter.com/petebankhead/status/1598012187833794560?s=20&t=zxPKZT2TuArSgGcuXPdzQw
For example, here's a pre-trained object detection model from DJL's PyTorch model zoo running through QuPath. Results are converted to QuPath-friendly annotations & classifications.
What used to be horribly complicated in QuPath, now just takes a few lines of code in a script
What used to be horribly complicated in QuPath, now just takes a few lines of code in a script

Here's another example using object detection and semantic segmentation models.
Running through QuPath means it's easy to restrict the segmentation to just part of the image, further refine the ROIs, adjust the input resolution, or export the results.

Running through QuPath means it's easy to restrict the segmentation to just part of the image, further refine the ROIs, adjust the input resolution, or export the results.


And here's a style transfer model, with the output added as a QuPath overlay.
Of course you can use your own models as well - it just needs to be in a DJL-friendly form.
Full code & explanations are at qupath.readthedocs.io/en/0.4/docs/de…
Of course you can use your own models as well - it just needs to be in a DJL-friendly form.
Full code & explanations are at qupath.readthedocs.io/en/0.4/docs/de…
But what if you want some more bioimage-related models - ideally ones you don't have to train yourself?
The Bioimage Model Zoo people have been working on that.
The Bioimage Model Zoo people have been working on that.
https://twitter.com/ilastik_team/status/1534822479927164929?s=20&t=5OKIp4Xaaket6DSd7qBtiw
QuPath v0.4.0 has initial support for a subset of models from the model zoo at bioimage.io
It's still early & not everything works as well as it should, but it's already possible to convert a few segmentation models into QuPath pixel classifiers
It's still early & not everything works as well as it should, but it's already possible to convert a few segmentation models into QuPath pixel classifiers
Once a deep learning model gets wrapped up into a pixel classifier, it behaves like any pixel classifier in QuPath - supporting measurements, creating objects, applying classifications etc.
There is more info at qupath.readthedocs.io/en/0.4/docs/de…
There is more info at qupath.readthedocs.io/en/0.4/docs/de…
• • •
Missing some Tweet in this thread? You can try to
force a refresh