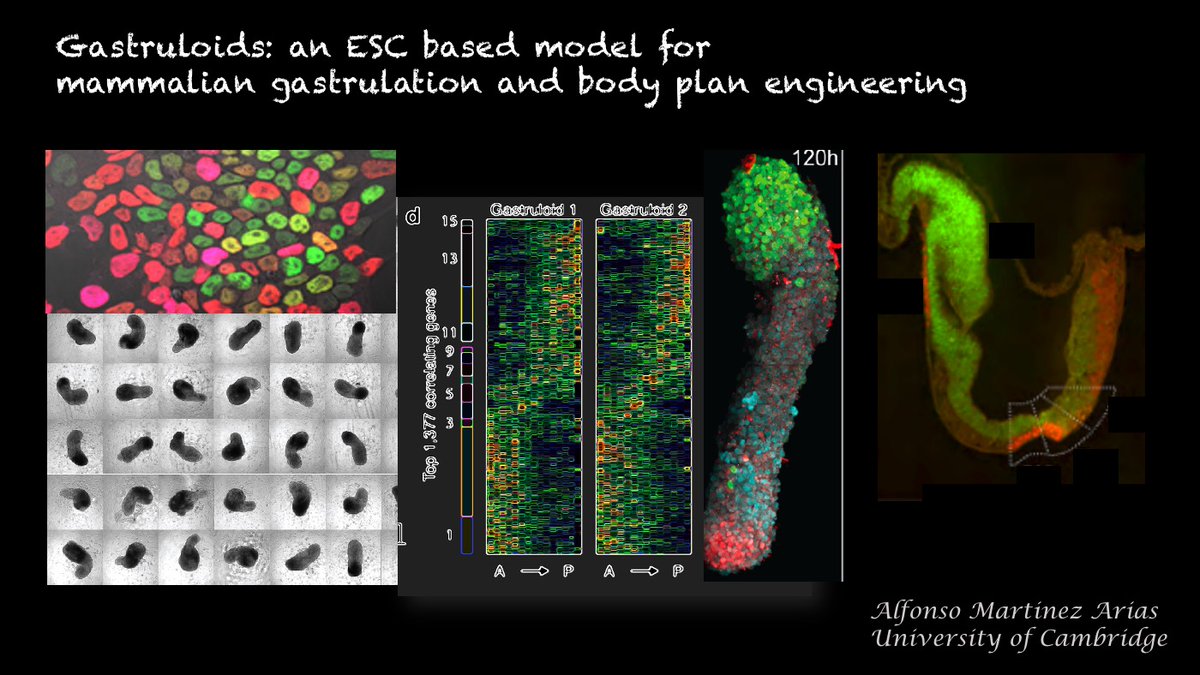
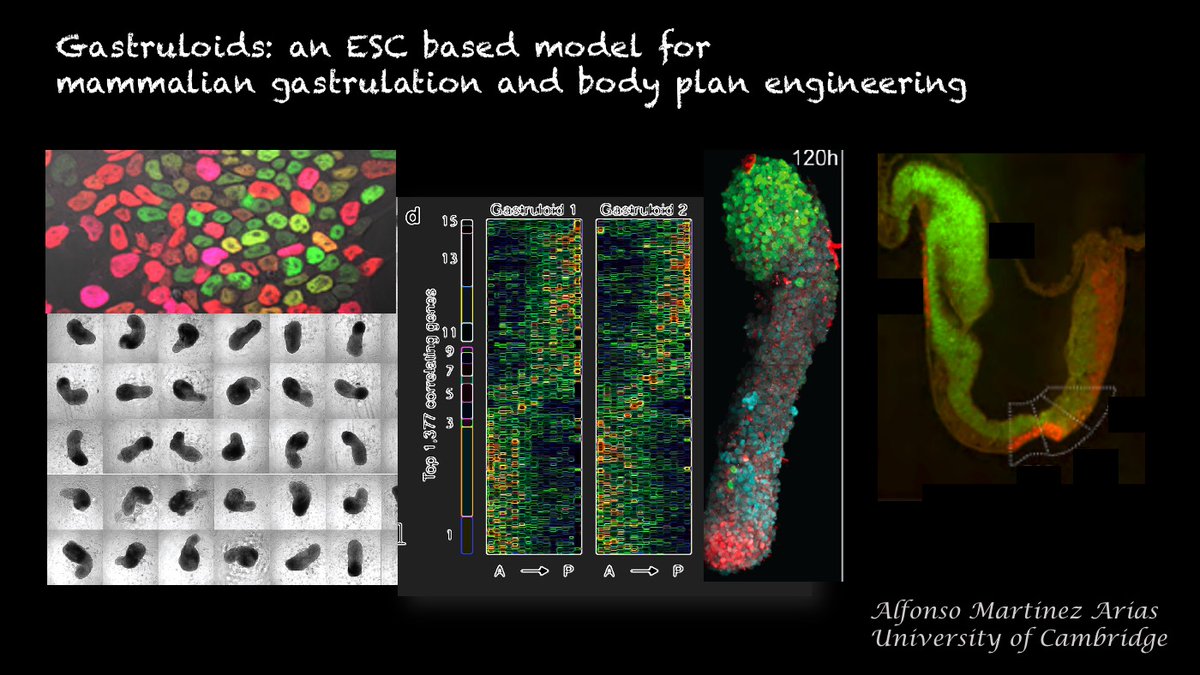
It is difficult to understand why @Nature publishes papers like this nature.com/articles/s4158… whose title promises much but which in the end is, simply, a data set with little insight & a number of misleading statements.
Perhaps the most valuable figures, which are not discussed in sufficient detail, are 2d and 4b where there we can see some spatial expression. Rather than genes that are the same, it would have been good to see differences.
Overall the technical work is good, probably useful, but the study is a superficial and misleading annotation of the data set to cell types: CDX1 and OTX2 as markers for Definitive Endoderm? HOX as a marker for the node?.
Somitogenesis and neural differentiation are, in general, well annotated though, there's a lot of data out, and as basically all they do is interpret their data on what is known from mouse, it is difficult to know if there are any differences.
Every assignment to the node should be questioned e.g. to pick HES4 as a halmark for ‘anterior mesoderm within the organizer” (?) is, at the very least enigmatic, not to mention the following sentence and the reference to its control of proliferation with 2 irrelevant references.
I could go on but it must be disturbing for Dev Biologists doing functional studies & proper #embryology to see this jumble of genes and misinformation.
Given the need to understand human #gastrulation, we need to make sure that annotations and interpretations of related data are carefully presented and discussed not to mislead in the future.
It seems to me that @Nature should start a section, preferably on line, w/ ½ page advertisement on the main journal, for these data sets. Studies like this don’t help the field. Unless they've more content, they shouldn't have this prominence w/o a justification. #disappointed
• • •
Missing some Tweet in this thread? You can try to
force a refresh