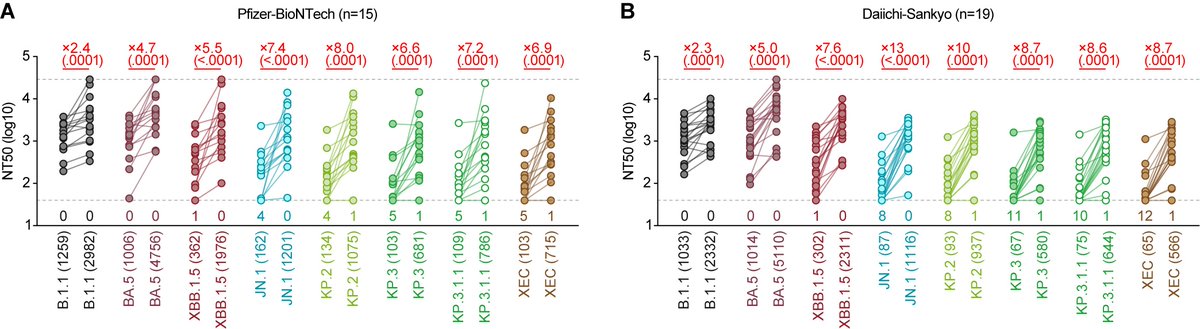
BREAKING🔔 We introduce the latest work from G2P-Japan🇯🇵 focusing on the virological characteristics of new SARS-CoV-2 lineage of concern, XBB.1.5 (aka #Kraken), which is rapidly spreading in the US🇺🇸 Please RT! Preprint/peer-review paper will come soon🔥 1/
XBB.1.5 = XBB.1 + #F486P in spike. We also found that a part of XBB.1.5 has reversion mutation in Y144del (=XBB.1.5 + #ins144Y) in spike. Because #Y144del evades antiviral immunity and decreases pseudovirus infectivity, its reversion might be also critical. 2/ 



First, a brilliant young talent, @jampei2, showed that the relative basic reproduction number (Re) of XBB.1.5 is >1.2-fold greater than the parental XBB.1 (❗️). Importantly the "1.2-fold difference" is similar to the differences between BA.1 versus BA.2 and BA.2 versus BA.5. 3/ 

We then showed that the KD value of XBB.1.5 spike is greatly lower than that of XBB.1 spike, meaning that XBB.1.5 spike exhibits very strong affinity to human ACE2, which is attributed to #F486P mutation. 4/ 

Experiments using pseudovirues also showed the ~3-fold increase of infectivity by #F486P mutation (XBB.1 << XBB.1.5). On the other hand, #ins144Y increased the infectivity of XBB.1 (XBB.1 < XBB.1+ins144Y) but did not increase that of XBB.1.5 (XBB.1.5 = XBB.1.5+ins144Y). 5/ 

Neutralization assay showed that XBB.1.5 is robustly (41-fold vs B.1.1, 20-fold vs BA.2) resistant to BA.2 breakthru infection sera. XBB.1.5 is also severely (32-fold vs B.1.1, 9.5-fold vs BA.5) resistant to BA.5 breakthru infection sera. 6/ 
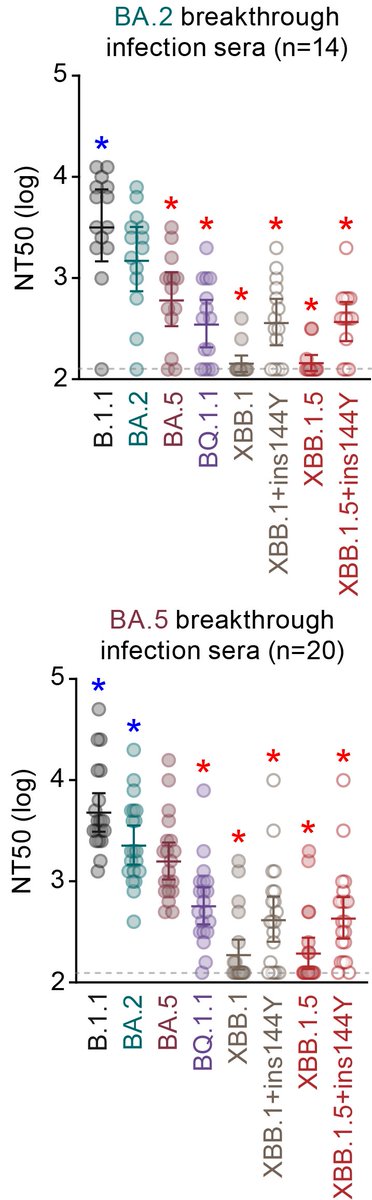
In sum, our results suggest:
XBB.1.5 = XBB.1 + F486P = profound immune resistance + augmented ACE2 binding affinity → increased transmissibility. 7/7
XBB.1.5 = XBB.1 + F486P = profound immune resistance + augmented ACE2 binding affinity → increased transmissibility. 7/7
• • •
Missing some Tweet in this thread? You can try to
force a refresh