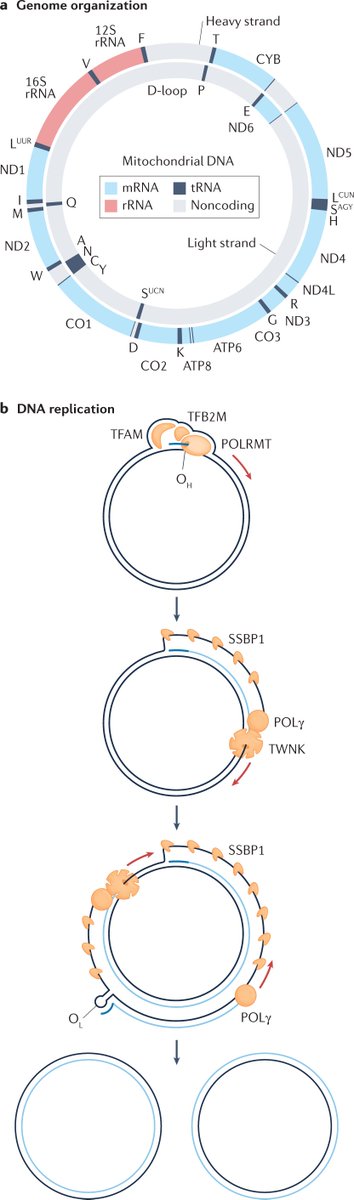
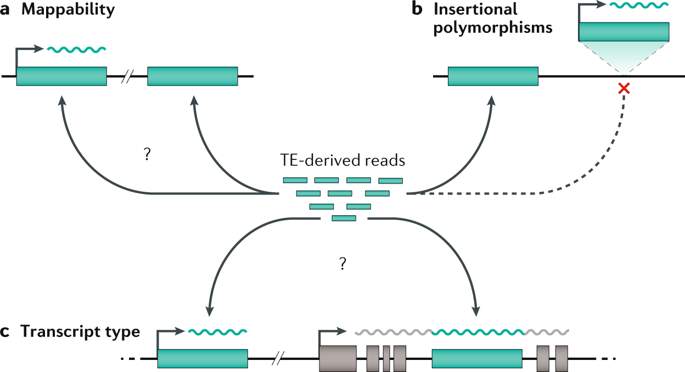
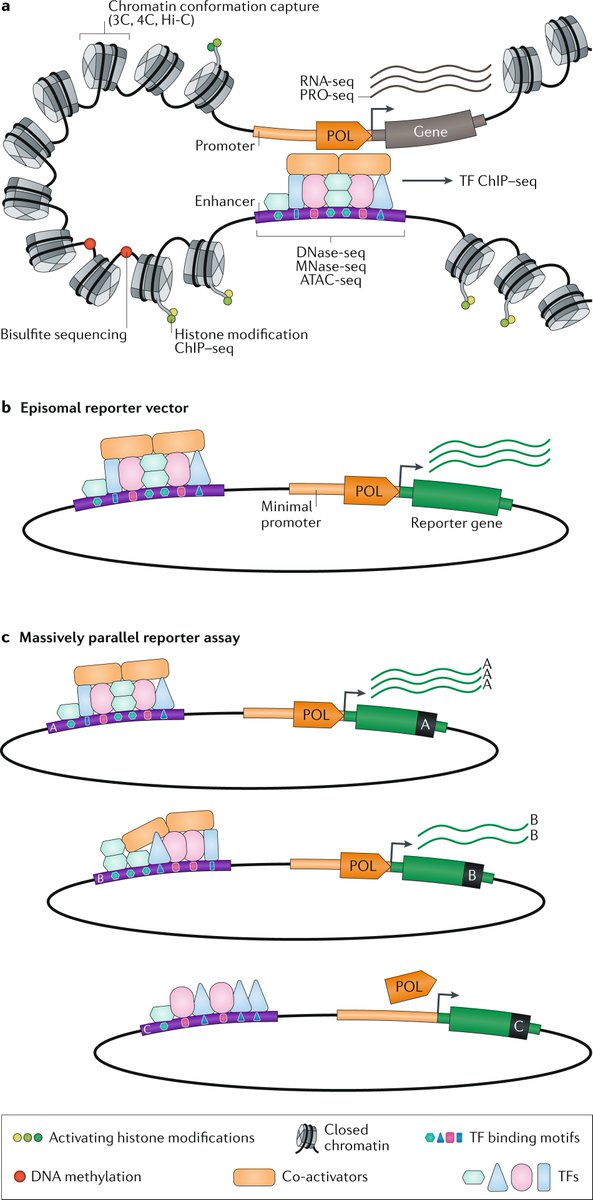
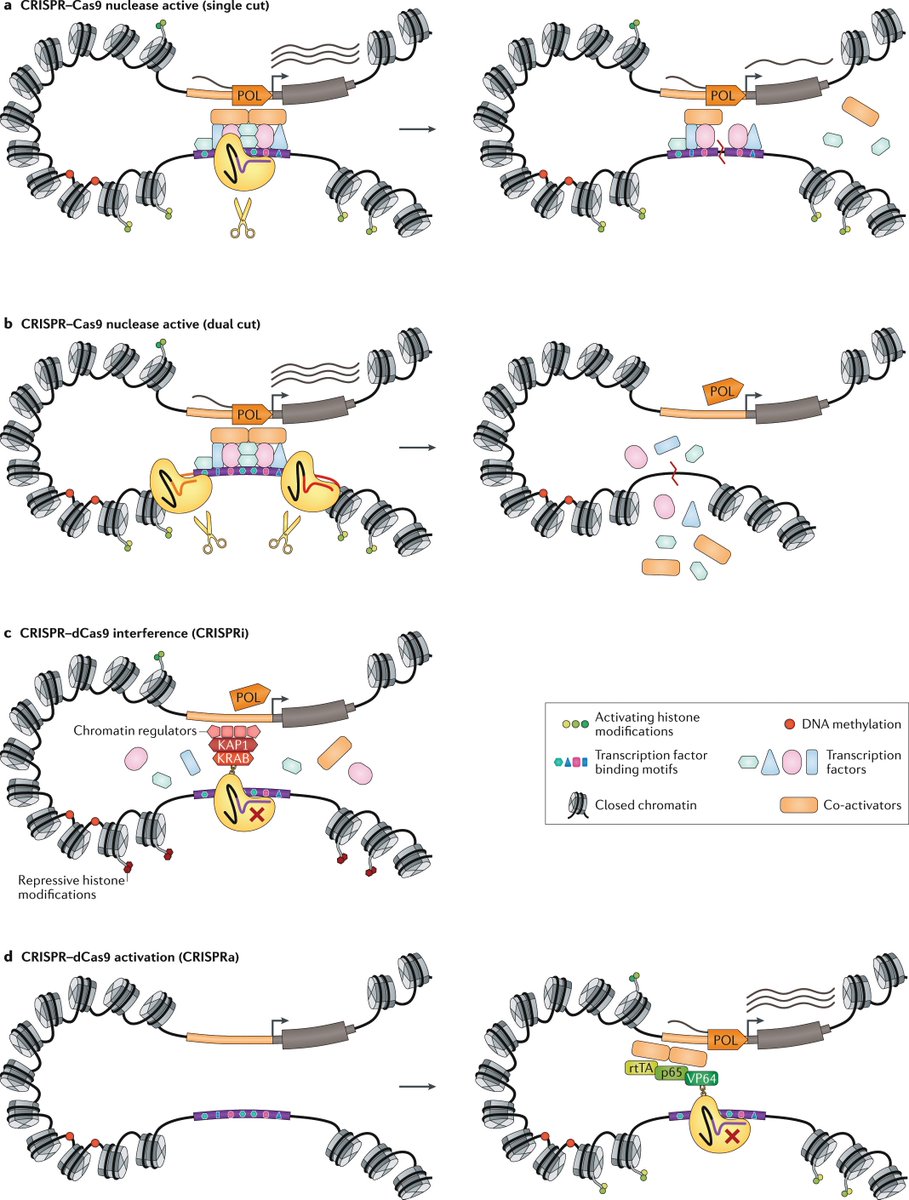
Human-specific genetics: new tools to explore the molecular and cellular basis of human evolution go.nature.com/3JLDI2I #Review by @Lab_Pollen, @KlkUmut, Craig B. Lowe & @GrayCampLab
@UCSF @UniBasel_en @DukeU
@UCSF @UniBasel_en @DukeU
In this Review, the authors discuss our latest understanding of evolutionary genetic changes that are specific to humans, which might endow uniquely human traits and capabilities 

They describe how new cellular and molecular approaches are helping to decipher the functional implications of these human-specific changes 

Free to read here: rdcu.be/c4Pno
• • •
Missing some Tweet in this thread? You can try to
force a refresh