As we see videos in social media of the first $PACB PacBio Revio to enter the @broadinstitute , with the promise of the long-read $1000/genome in place, what does it look like today to set up a Next-Generation Sequencing factory/institute?
https://twitter.com/AlbertVilella/status/1624465272496168965
There have been many of these institutes come and go, and probably the @broadinstitute (Cambridge, US) and the @sangerinstitute (Cambridge, UK) are the two referents in historical terms and in their magnitude of achievements in #genomics. So if we take them as an example, what do
we learn from them?
1) They were successful at being early adopters of large-scale Sanger Sequencing.
2) They were very well funded from the get-go, and contributed in large part to the success of sequencing the reference #human #genome in 2000-2001.
1) They were successful at being early adopters of large-scale Sanger Sequencing.
2) They were very well funded from the get-go, and contributed in large part to the success of sequencing the reference #human #genome in 2000-2001.
3) They were early adopters of NGS, first with 454 and Solexa, then with the successive iterations of ABI SOLID, $ILMN Illumina and $PACB PacBio instruments.
4) The exomes and 30x genome factories using short-reads were designed and implemented by them.
4) The exomes and 30x genome factories using short-reads were designed and implemented by them.
Is there anything else we can learn from them?
We can say that they have been adopters of $ONTTF Oxford @nanopore but not to the same extend to $PACB PacBio technology. One of the big ponchos at one of the institutes really didn't like ONT's technology,
We can say that they have been adopters of $ONTTF Oxford @nanopore but not to the same extend to $PACB PacBio technology. One of the big ponchos at one of the institutes really didn't like ONT's technology,
but he is not in there anymore, so their influence is waning.
What can we learn from more recent genomic institutes and the trends they follow? I'll take as an example one of the recently established institutes in the Middle East, and many of you will correctly guess which one
What can we learn from more recent genomic institutes and the trends they follow? I'll take as an example one of the recently established institutes in the Middle East, and many of you will correctly guess which one
I am referring to without naming it.
1) They are investing in MGI Tech's DNBSEQ factory sequencers for the short-read side of things. Now that the DNBSEQ T20x2 is available, they'll soon have the equipment to produce $100/genomes,
1) They are investing in MGI Tech's DNBSEQ factory sequencers for the short-read side of things. Now that the DNBSEQ T20x2 is available, they'll soon have the equipment to produce $100/genomes,
outcompeting both the @broadinstitute and the @sangerinstitute in their ability of producing short-read data at $1/Gb.
2) They have the largest (public) installation of Oxford @nanopore PromethION instruments under one roof. They don't have any @PacBio instruments that I know.
2) They have the largest (public) installation of Oxford @nanopore PromethION instruments under one roof. They don't have any @PacBio instruments that I know.
What does the combination of (1) and (2) mean for this newly establish genomic institute?
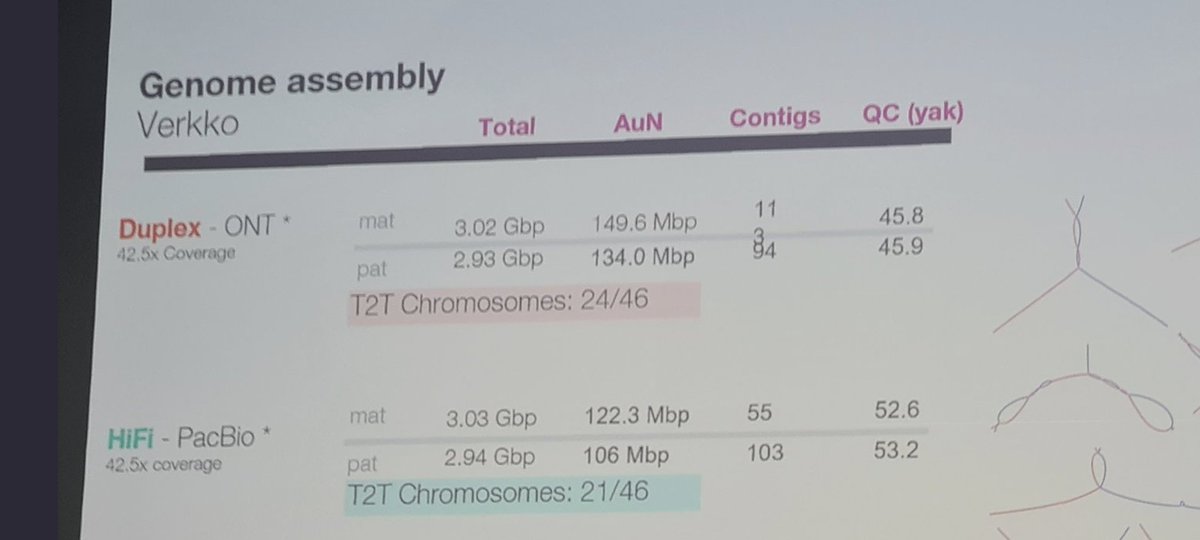
Let's forget about high duplex Q30+ reads for the moment, but just concentrate on the Ultralong Oxford @nanopore reads,
Let's forget about high duplex Q30+ reads for the moment, but just concentrate on the Ultralong Oxford @nanopore reads,
a technology that is available directly from the nanoporetech.com shop, and has shown to produce very good results in the T2T benchmarks which were done recently.
What does the ULR tech do combined with $1/Gb short-reads for a genomic factory?
What does the ULR tech do combined with $1/Gb short-reads for a genomic factory?
The T2T project has already answered many of the connotations of these two technologies, Ultralong reads and cheap short-reads, but it is worth mentioning what these are for the post #AGBT23 era.
When sequencing a personal human genome, if done only with short-reads, there are parts of the genome that are inaccessible for certain type of diagnostics. Some put the mark of the accessible short-genome at 80% of the total, some put it somewhere else.
Some of these difficult areas contain genes like FXN, FMR1, DMD, CYP21A2, HBA1,HBA2, SMN1, among others, which are important for many areas in diagnostics.
What difference does it make to obtain a complete personal genome with tech like ULRs plus short-reads?
What difference does it make to obtain a complete personal genome with tech like ULRs plus short-reads?
If done with the combination of these two technologies, these effectively allow for a fully sequenced personal genome without any inaccessible areas, with high precision and recall in both SNPs and Indels, and also structural variants and copy number alterations.
Once you have the new reference personal human genome for a given individual, this can be used as an Electronic Health Record, some would say the most important EHR for that individual, and refer to that record over and over along the medical life of that person.
If the personal genome was done for a new-born person, it'll be available for the first 5 years of their life, which are very important developmental years, and also a period of time of susceptibility to blood cancers, such as leukaemia.
Some of the drugs and biologics used for childhood cancers have successfully increased the survival rates in the last 10-15 years, and being able to use Liquid Biopsy with cfDNA NGS means we can monitor the remission of such cancers after they have been treated.
Some cancers, like testicular cancer, can also appear in the early years of life, so people in the age range of 18-23 yo will undergo diagnostic and treatment for those. Here is another area where Liquid Biopsy cfDNA NGS can help, both for diagnostics and checking the
progression of the treatment and remission of the cancer. Epigenomic profiling of cfDNA with NGS can achieve higher levels of diagnostic success of small pockets of cancer cells that are near impossible to detect via somatic mutation profiling. This will revolutionise the field.
The biggest difference that NGS will make to human health is in the early diagnostic of cancer in screening the blood of people in the 45+ age range. Again it will be epigenomic profiling of cfDNA with NGS, both 5mC and 5hmC, that will change the way we diagnose and treat cancer.
Oxford @nanopore has already shown to be able to differentiate the unmethylated CpGs from 5mC and 5hmC CpGs, and do it for the fragment ranges in cfDNA, the length of which contains important information about which molecules come from healthy cells or cancerous cells.
Although @PacBio has shown to be able to do 5mC natively, they haven't shown any data for 5hmC, and the library prep needs to concatenate the short fragments in cfDNA into longer circular fragments before being able to put them in a PacBio sequencer.
The situation is similar for short-reads sequencers: one needs to pre-treat the short-fragments of DNA with either chemical of enzymatic modifying steps to be able to differentiate the As, Ts, Gs, and Cs (in CpG context), from 5mCs. Only one or two technologies can assay 5mC+5hmC
in one go (I'll let you Google which ones, as I wouldn't be accused of peddling my own you-know-what). So the ways in which one can assay ACGT+5mC+5hmC are limited, and they are very important in cfDNA cancer screening.
Finally I should mention that all this recursive screening of cfDNA from the age of 45 or older will happen first for cancer screening, but the same technology can be used for other conditions that have had less attention and investment until now.
Cardiovascular and metabolic diseases is one of such areas. We are here in the enviable position of having been able to treat a large fraction of the aging population with statins, which have been cheap, available and well understood for years now.
Diseases of the brain and the central nervous system are another area where sequencing cfDNA with NGS will become important. Some work has already been done in cases like Multiple Sclerosis (MS), and more will come from other neurodegenerative diseases that have high prevalence.
Another area where NGS will have an importance in human health is Spatial Biology. This brings back the original biopsy, rather than the liquid biopsy, to the fore, and in this space, NGS and #AI will be able to do more than the cruder methods of staining biopsies currently used.
The #pathologists following this account shouldn't fear for their livelihoods yet though! Rather than replacing them, this will be, in my opinion, an important weapon in their arsenal to diagnose and guide treatment of disease.
Another consequence of more affordable and available NGS will be in #Antibody #Discovery. Tools such as single cell assays combined with NGS have turbo-charged the ability of designing antibodies to treat diseases for which small molecules aren't covering all needs.
But before you make a therapeutic antibody to target a protein that is causing a disease, you need to know which protein to target. How do you identify such targets? Yes, you guessed it correctly, with NGS! Companies like $REGN Regeneron are spending a lot of time and effort in
applying NGS to target identification and validation. But there will be disruptors to this area which are themselves targeting Regeneron and their lucrative market share.
So all in all, if NGS is so useful, why aren't there more of these "genomic centers" I keep talking about?
Well, exactly! There are in fact many of them, as long as you adapt the definition of a "genomic center" to what the post #AGBT23 era signifies.
Well, exactly! There are in fact many of them, as long as you adapt the definition of a "genomic center" to what the post #AGBT23 era signifies.
So what do you need at its minimum for your own "genome center"? Can you set up one in your garage? The answer is: yes, you can. The second question is: should you? The answer will most of the time be: "no", especially if it comes from your unfazed partner.
1) An Oxford @nanopore MinION starter kit will give you the first starting point for your own "genome center". Your now disbelieved partner will be glad to know you only spent a few thousand dollars and it all fits in a drawer in the garage that was full of junk anyway.
2) With some basic library prep equipment, a pipette and a few off the shelf kits, you will be able to do your own libraries and sequence them with your own sequencer.
But that's not the same as the @broadinstitute or the @sangerinstitute , is it?
But that's not the same as the @broadinstitute or the @sangerinstitute , is it?
3) If you want the $1/Gb price of short-reads or anywhere near that, your version 1 of your own genomic center won't give you that straight-away. But if you can isolate the DNA and maybe even prepare your own libraries, you can send it to a service supplier like NovoGene.
4) Once you start churning ULR Oxford @nanopore reads in your garage and receive the FASTQ files from NovoGene, you'll need to start analysing the data. Get yourself a tower computer, ideally with a 16GB VRAM GPU card from @nvidia, and install Linux on it.
There is plenty of help on how to get started in Bioinformatics, but the EBI training website is ebi.ac.uk/training/ a good place to start. Genomics isn't difficult. Even myself can do it!
5) You've started churning ONT ULR data in your garage and short-reads from an external supplier, and now that you've got the kick for it, you want to upgrade your rig. What's the next step? An Oxford @nanopore P2 Solo will set you back another $10K, but now you are churning
long-reads at a higher throughput and at a fraction of the cost of the original MinION starter kit, which you can now use with Flongle flowcells to validate your library preps, maybe also to find out which dog is pooing on your lawn (they already do that in Switzerland).
6) Given an ONT P2 Solo, MinION, Flongles and basic equipment for long-read and short-read library prep, what's the next step to increase the capabilities of a "genomic center". We will leave the garage analogy from here onwards as the costs will go up considerably from now on.
7) Buying 3 of the AVITI sequencers from Element Bio @ElemBio will give the newly formed genome center the ability of sequencing short-reads at $2/Gb, and reaching the thousands mark per year. The total OPEX will still be below $1M, including liquid handlers for automated preps.
An Oxford @nanopore PromethION P24 or P48 will give this newly formed "genome center" the ability to produce Ultra Long Reads at the same pace as the @ElemBio AVITIx3 instruments are producing short-reads. So now we have a system for T2T genomes in the thousands per year.
8) If churning short-reads at $2/Gb isn't good enough, you'll be able to upgrade your genomic center with an MGI Tech DNBSEQ-T7, and benefit from a better $1.5/Gb price. If you have enough space, you can install a DNBSEQ-T20x2 to generate $1/Gb short-reads.
If you haven't enough space, then you have until Q3 2023 to build your genomic center larger, as that will be the point where the $1/Gb price kicks in.
• • •
Missing some Tweet in this thread? You can try to
force a refresh