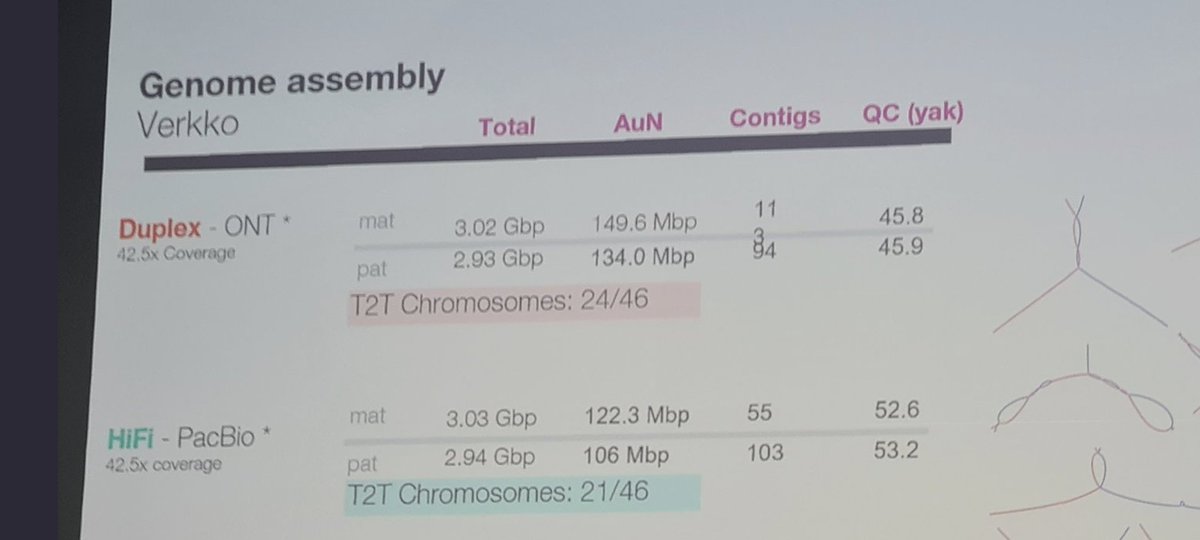
In #biotech #stocks news, today we review $TXG @10xGenomics , one of the companies in the #SingleCell and #SpatialBiology sectors.
10x Genomics is a biotechnology company that develops and markets solutions for omics analysis of single cell and spatial biology samples. The company offers several product lines:
The Chromium platform is used for single-cell gene expression analysis, immune profiling, and genome sequencing. It allows researchers to study gene expression at the single-cell level, which can provide insights into cell behavior and disease progression.
Competitors in this space include other companies offering single-cell analysis platforms, such as Fluidigm, Mission Bio, and BD Biosciences. As of late, a new breed of instrument-free companies have also appeared, including Parse Bio, Fluent Bio, BioSkryb and others.
Visium is a spatial gene expression analysis platform that allows researchers to map gene expression in intact tissues. This can provide information about cell types and their interactions in complex tissue environments. 10X is putting together the launch of their Visium HD.
Competitors in this space include companies like NanoString and Curio Bio. BGI Tech is also putting together a commercial product equivalent to all these NGS-based products for Spatial Biology.
Xenium is the third line of products that 10X has recent put in the market. This is an in-situ imaging device which instead of giving an NGS-based readout to a biopsy, it gives an imaging-based readout of either RNA or proteins present in the sample.
Competitors in this space include NanoString's CosMx, Vizgen MERSCOPE, Akoya Biosciences products, Resolve Bio Molecular Cartography, Single Technologies Theta, Veranome Bio, Lunaphore Technologies, Ultivue, Syncell and others.
More at bit.ly/scspatial-slid…
• • •
Missing some Tweet in this thread? You can try to
force a refresh