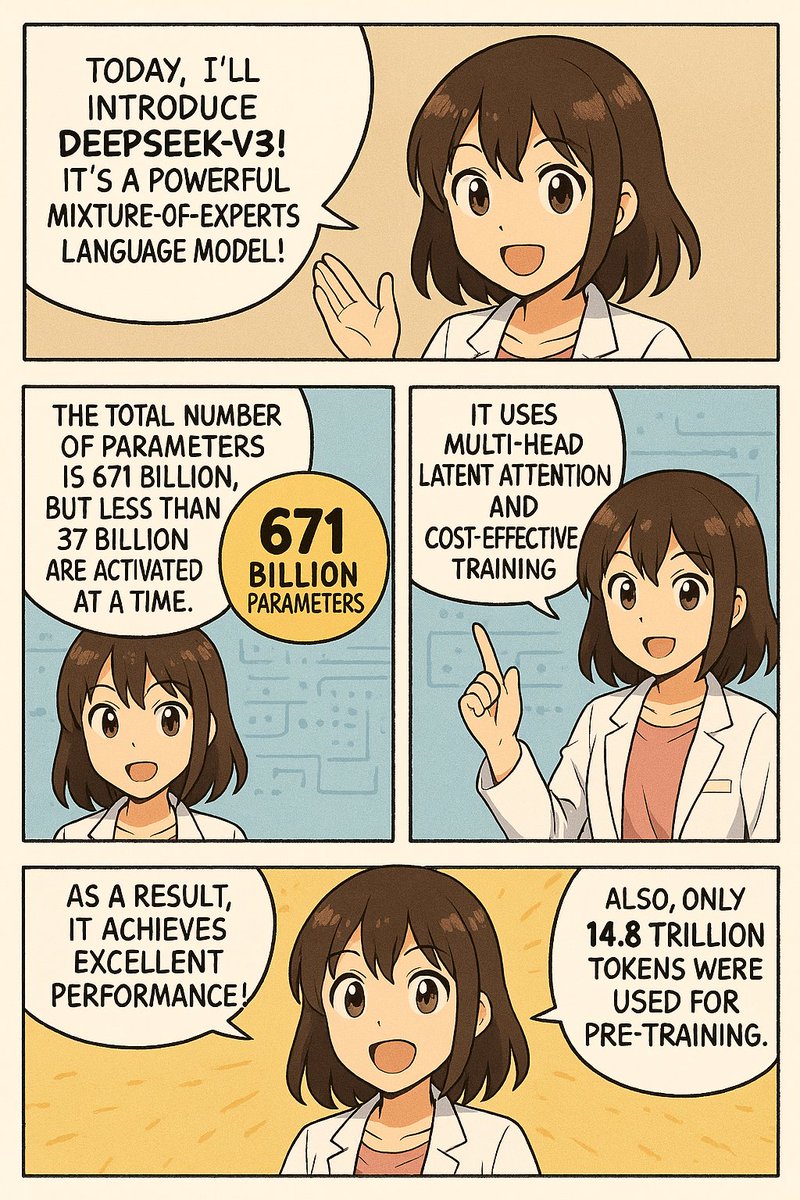
How does GPT-4 do in the medical domain?
I got to play around with its multimodal capabilities on some medical images!
Plus a recent Microsoft paper examined its text understanding and got SOTA results on USMLE medical exams!
A quick thread ↓
I got to play around with its multimodal capabilities on some medical images!
Plus a recent Microsoft paper examined its text understanding and got SOTA results on USMLE medical exams!
A quick thread ↓
As I showed earlier, I had the chance last week to play around with GPT-4's multimodal capabilities:
https://twitter.com/iScienceLuvr/status/1636479850214232064
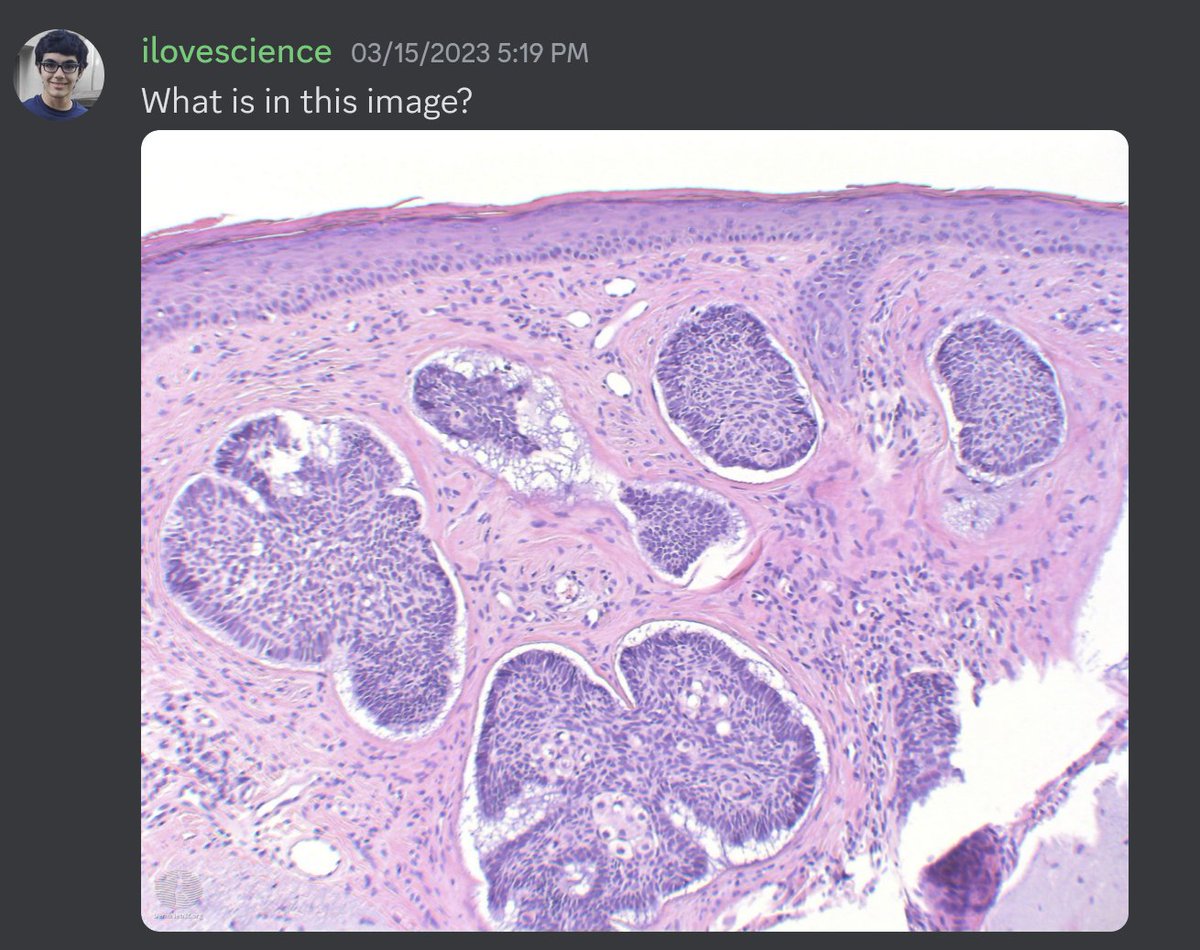
I also tried some medical images too! Here I started with some histopathology. I passed in an H&E image of prostate cancer and asked GPT-4 to describe it. It knew it was an H&E image of glandular tissue but was unable to identify it as low grade prostate cancer. 

Here I passed in an image of invasive lobular carcinoma with characteristic single file lines of tumor nuclei. It fails to notice this unfortunately not matter how hard I try. 





Here is an example of a glioblastoma (severe brain tumor). It has a characteristic feature again that suggests the glioblastoma diagnosis (pseudopalisading necrosis) but it fails to notice that. It does realize the presence of what looks like tumor nuclei. 

This image shows H&E of basal cell carcinoma (skin cancer). GPT-4 notices that it is of skin but cannot identify the pathology. 


Overall though, GPT-4 mostly refuses to provide anything similar to a diagnosis. Here is one such example with and X-ray image. 

My conclusion on the multimodal side is that GPT-4 is a impressive first step towards multimodal medical understanding, but its understanding right now is fairly rudimentary, and there is a lot of room to improve here.
On the text side of things, however, the situation is different. In a recent paper from Microsoft Research, "Capabilities of GPT-4 on Medical Challenge Problems", GPT-4 obtains SOTA on USMLEs (medical student exams), significantly outperforming GPT 3.5. 

One may worry the high performance is due to data contamination. Interestingly this paper performed a memorization analysis, and they didn't find any of the tested USMLE questions with their memorization detection (though it doesn't 100% confirm no memorization). 

Plus the USMLE material is behind paywall and probably unlikely to be in the GPT4 training set anyway. 

Overall, seems the medical understanding of text-only GPT-4 is significantly improved & multimodal GPT-4 has rudimentary understanding.
Many more experiments should be done to study GPT-4's medical knowledge/reasoning. Some previous studies using GPT-3 concluded domain/task-specific fine-tuned model are better, and I wonder if the conclusion changes now with GPT-4.
#MedTwitter #PathTwitter
#MedTwitter #PathTwitter
If you like this thread, please share!
Consider following me for AI-related content! → @iScienceLuvr
Consider following me for AI-related content! → @iScienceLuvr
• • •
Missing some Tweet in this thread? You can try to
force a refresh