Only a matter of time before a paper formalized this exercise:
Automated #scRNAseq cell type annotation with GPT4, evaluated across five datasets, 100s of tissues & cell types, human and mouse.
A🧵below with my thoughts on how such tools will change how #Bioinformatics is done.
Automated #scRNAseq cell type annotation with GPT4, evaluated across five datasets, 100s of tissues & cell types, human and mouse.
A🧵below with my thoughts on how such tools will change how #Bioinformatics is done.

I'll start with a quick summary of the paper, such that we're all on the same page.
(The paper is also a super quick read, literally only 3 pages of text, among which 1 is GPT prompts).
Here's the link to the preprint
biorxiv.org/content/10.110…
(The paper is also a super quick read, literally only 3 pages of text, among which 1 is GPT prompts).
Here's the link to the preprint
biorxiv.org/content/10.110…
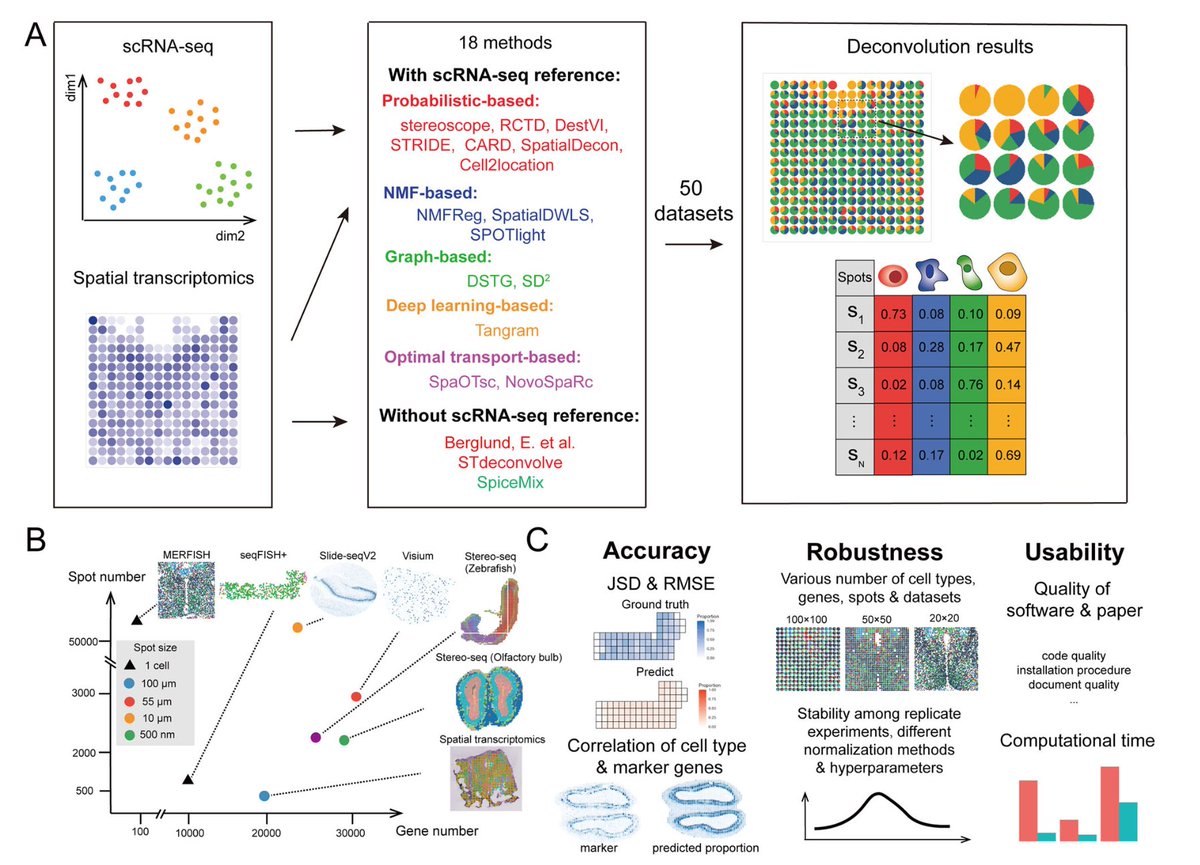
The paper looked at 4 already-annotated public datasets: Azimuth, Human Cell Atlas, Human Cell Landscape, Mouse Cell Atlas.
Differentially Expressed Genes characterizing every cluster (DEGe) in these studies were generally available with the publications & were also downloaded.
Differentially Expressed Genes characterizing every cluster (DEGe) in these studies were generally available with the publications & were also downloaded.
In addition to these 4 sets of DEGs, the authors also downloaded a large set of marker genes from the Human Cell Atlas.
GPT4 was prompted (with basic prompts) to identify cell types in these 5 scenarios, given some of the top DEGs in each list.

GPT4 was prompted (with basic prompts) to identify cell types in these 5 scenarios, given some of the top DEGs in each list.


Assessment:
“fully match” if GPT4 & original annotation is the same cell type;
“partially match” if the two annotations are similar, but distinct cell types (e.g. monocyte & macrophage);
“mismatch” if the two annotations refer to different types (e.g. T cell & macrophage).
“fully match” if GPT4 & original annotation is the same cell type;
“partially match” if the two annotations are similar, but distinct cell types (e.g. monocyte & macrophage);
“mismatch” if the two annotations refer to different types (e.g. T cell & macrophage).

GPT4's performance in matching the cell type annotation in the original publications is quite impressive.
Of course some tissues are better than others, but given the little effort involved, it certainly is able to provide a good intuition about the analyzed data.
Of course some tissues are better than others, but given the little effort involved, it certainly is able to provide a good intuition about the analyzed data.
The authors note that GPT4 is in best agreement with the original annotations when using only top 10 differential genes, and using more may reduce agreement.
This is an insightful observation, which essentially means that GPT4 is a great "literature summarizer" for what is known
This is an insightful observation, which essentially means that GPT4 is a great "literature summarizer" for what is known
This also means that GPT4 is great for annotations heavily relying on canonical marker genes.
So, if your pipeline involves lots of comparing with literature/googling/extracting expert info, as opposed to just using algorithms on DEGs, then GPT4 can be very helpful.
So, if your pipeline involves lots of comparing with literature/googling/extracting expert info, as opposed to just using algorithms on DEGs, then GPT4 can be very helpful.
Another straight-forward observation is that GPT4 does best when the cell type is more homogeneous, and worst when it is a heterogeneous mixture (see stroma).
(totally expected)
Still, there's prompting tricks that one can do to increase accuracy.
(totally expected)
Still, there's prompting tricks that one can do to increase accuracy.
Now, what does this all mean?
1. We need to understand that cell type assignment is the perfect *type of problem* on which chatGPT excels
Why?
Ultimately, most cell assignments boil down to expert knowledge,which is nothing more than mirroring literature in a structured manner
1. We need to understand that cell type assignment is the perfect *type of problem* on which chatGPT excels
Why?
Ultimately, most cell assignments boil down to expert knowledge,which is nothing more than mirroring literature in a structured manner
All existing algorithms work by assessing similarities with existing databases/datasets, so in the end we need to choose among multiple ways to do the same thing.
Chatting with GPT4 happens to be a super quick way to get this task done, in a pretty reasonable manner.
Chatting with GPT4 happens to be a super quick way to get this task done, in a pretty reasonable manner.
2. In the near future, GPT-like models will be able to *reliably* automate most of bioinformatics pipeline running and interpretation.
For many Bioinformaticians and Data Scientists, this represents today a core part of the job.
For many Bioinformaticians and Data Scientists, this represents today a core part of the job.
- Writing & debugging code
- Running entire pipelines
- Interpreting results
- Creating reports
- Creating presentations based on results & reports
All these tasks can now be done in a fraction of the time that they used to occupy.
And the efficiency is quickly going up.
- Running entire pipelines
- Interpreting results
- Creating reports
- Creating presentations based on results & reports
All these tasks can now be done in a fraction of the time that they used to occupy.
And the efficiency is quickly going up.
This means more time for bioinformaticians to focus on higher-level tasks, for ex:
- understand the underlying biology
- structure & formulate questions
- learn about the reasoning behind the methods used (why is a method better than others, what are the limitations)
- understand the underlying biology
- structure & formulate questions
- learn about the reasoning behind the methods used (why is a method better than others, what are the limitations)
Similarly for more advanced algorithms (such as cell type assignment):
instead of getting lost for weeks/months in the weeds of installing/running/debugging such algorithms, chatGPT-like tools will be able to provide reasonable answers in literally 5 minutes.
instead of getting lost for weeks/months in the weeds of installing/running/debugging such algorithms, chatGPT-like tools will be able to provide reasonable answers in literally 5 minutes.
Of course, such results will likely be an initial attempt to solve the task, requiring further curation/inspection, and sometimes redoing.
But the time saved throughout the process is real, massive, and most importantly, it compounds (time is saved in a cascade of tasks).
But the time saved throughout the process is real, massive, and most importantly, it compounds (time is saved in a cascade of tasks).
3. For people who still think “how can I trust this, billions of parameters, this is not interpretable, this is not perfect”
REMEMBER
*none* of the algorithms out there (incl. expert intuition for e.g. cell assignment) is 100% perfect
perfect doesn’t usually exist in this game
REMEMBER
*none* of the algorithms out there (incl. expert intuition for e.g. cell assignment) is 100% perfect
perfect doesn’t usually exist in this game
4. None of this is to say that Bioinformaticians won't have an important role in driving the future biomed research.
On the contrary, the role of data & models is now more important than ever.
But biomedical science will overall move towards more & more interdisciplinarity.
On the contrary, the role of data & models is now more important than ever.
But biomedical science will overall move towards more & more interdisciplinarity.
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter