
Assistant prof at @StanfordMed. Interested in aneuploidy, mitotic kinases, cancer therapeutics, and drug development. Co-founder x2.
2 subscribers
How to get URL link on X (Twitter) App


https://twitter.com/biorxiv_cancer/status/1995323982615093495The drug is called PAC-1. It was initially developed to target cancer cells by activating the executioner caspases. But, we generated CASP3/6/7 triple-knockouts and it still eliminated cancer cells, demonstrating that it must have some other target.



 My top picks: Horwich/Hartl for their work on chaperone-mediated protein folding. Their discoveries changed how we think about protein structure and has had significant ramifications for our understanding of neurodegenerative diseases.
My top picks: Horwich/Hartl for their work on chaperone-mediated protein folding. Their discoveries changed how we think about protein structure and has had significant ramifications for our understanding of neurodegenerative diseases.

 To back up, I have a longstanding interest in understanding the trajectories of academic careers and uncovering “hidden” factors that influence success. Some of my published work on this topic:
To back up, I have a longstanding interest in understanding the trajectories of academic careers and uncovering “hidden” factors that influence success. Some of my published work on this topic:






 According to this data, "Nature" is not actually the most selective journal. Nature Med, Cancer, and Human Behavior all have lower acceptance rates.
According to this data, "Nature" is not actually the most selective journal. Nature Med, Cancer, and Human Behavior all have lower acceptance rates.





 We conducted a comprehensive analysis of genetic, epigenetic, and transcriptional features linked with patient outcomes across 10,000 patients and 32 cancer types.
We conducted a comprehensive analysis of genetic, epigenetic, and transcriptional features linked with patient outcomes across 10,000 patients and 32 cancer types.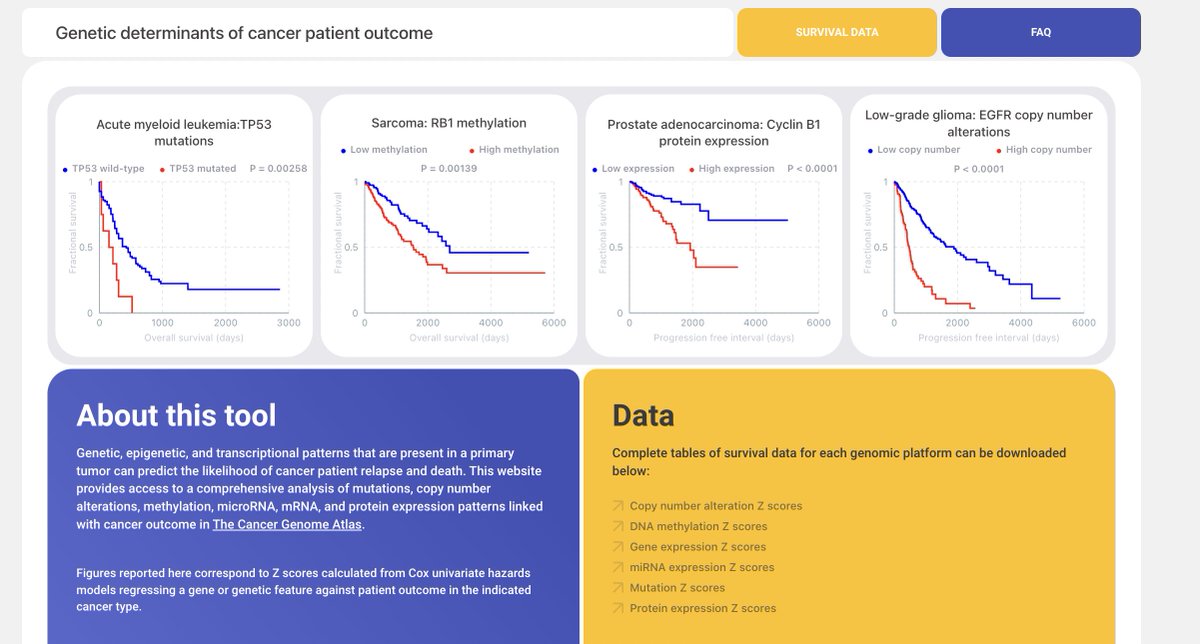

https://twitter.com/biorxivpreprint/status/1399950911573737474We used every type of data collected by TCGA (RNASeq, CNAs, methylation, mutation, protein expression, and miRNASeq) to generate survival models for each individual gene across 10,884 cancer patients. In total, we produced more than 3,000,000 Cox models for 33 cancer types.


 Some background: one of the best ways to collect real-world evidence of discrimination is through name-swapping "audit" studies. In these experiments, people are presented with job applications, resumes, mortgage applications, etc., that are identical except for the name…
Some background: one of the best ways to collect real-world evidence of discrimination is through name-swapping "audit" studies. In these experiments, people are presented with job applications, resumes, mortgage applications, etc., that are identical except for the name…

 To back up - my appointment at CSHL let me run a lab without doing a postdoc, so I never had the experience of applying for these grants. To help out my current postdocs, I wanted to make up for my lack of experience by doing some research.
To back up - my appointment at CSHL let me run a lab without doing a postdoc, so I never had the experience of applying for these grants. To help out my current postdocs, I wanted to make up for my lack of experience by doing some research.
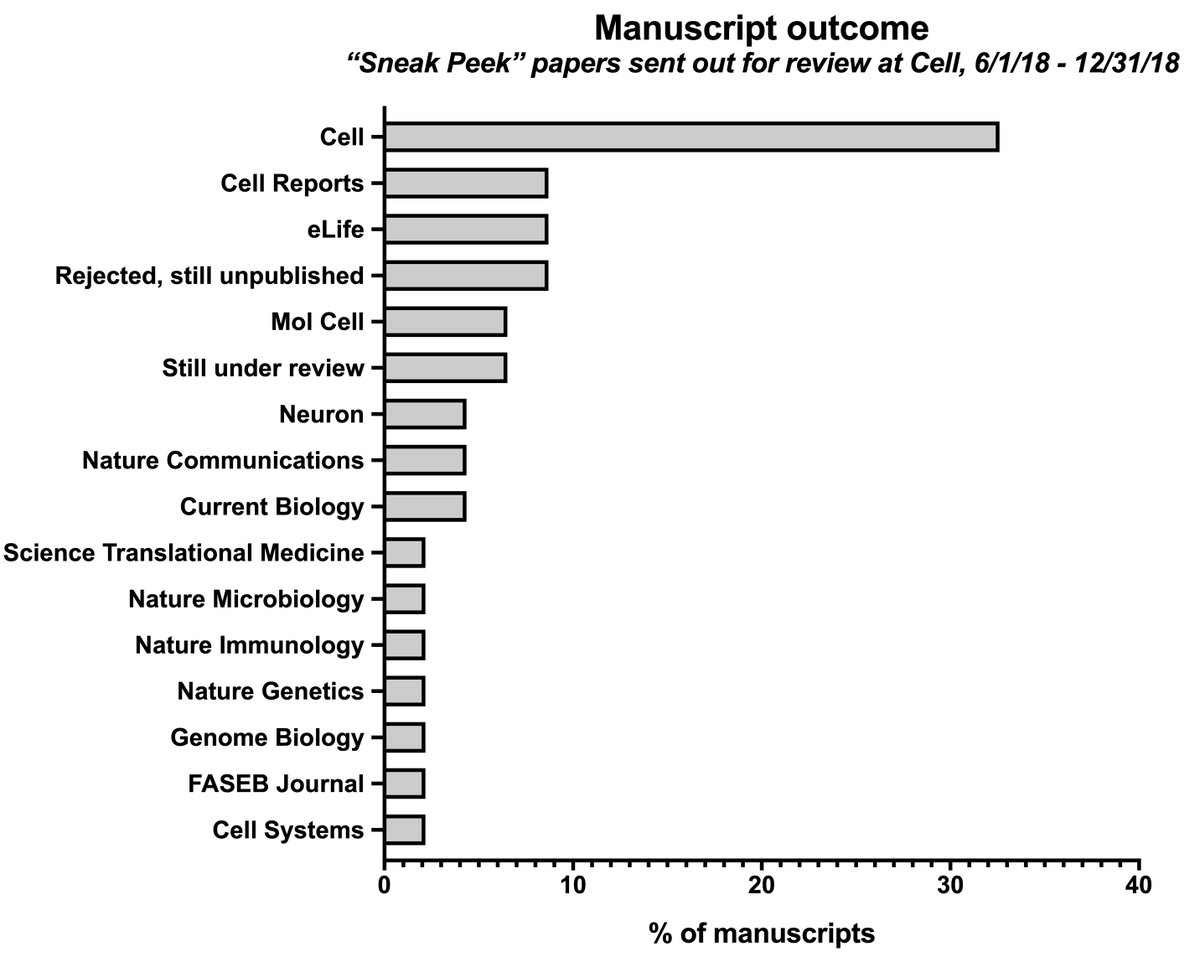

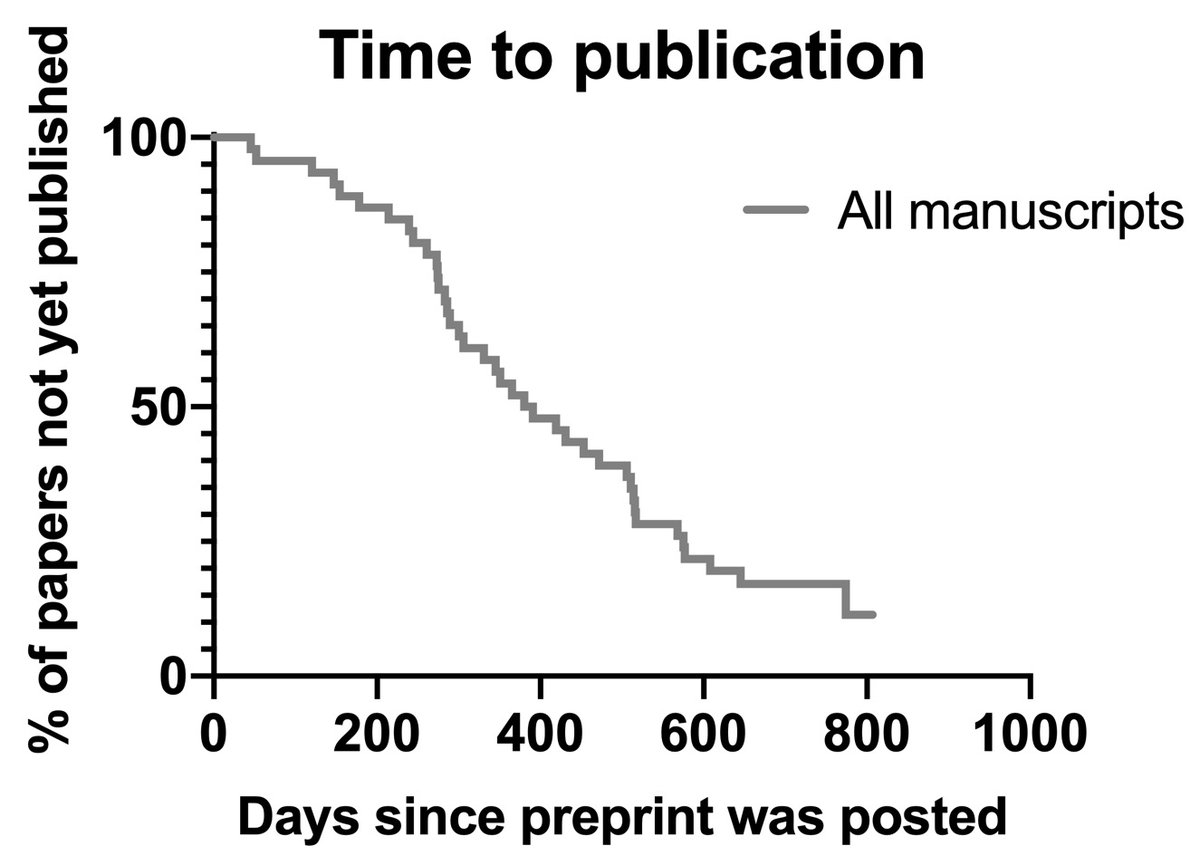 To back up: in 2018, Cell started the “Sneak Peek” program, in which authors had the option of posting a preprint of their manuscript if it was sent out for review by a Cell-family journal. cell.com/sneakpeek
To back up: in 2018, Cell started the “Sneak Peek” program, in which authors had the option of posting a preprint of their manuscript if it was sent out for review by a Cell-family journal. cell.com/sneakpeek

 A few thoughts: Wei recovered ACE2 as their #1 hit, which is strong evidence in favor of the biological validity of their screen.
A few thoughts: Wei recovered ACE2 as their #1 hit, which is strong evidence in favor of the biological validity of their screen.

 Methods: faculty candidate seminars were found on public departmental websites. Pub records were acquired from Google Scholar or Pubmed, and K99's were found on NIH Reporter. I did my best to summarize research topics by reading the abstracts of a candidate's recent papers.
Methods: faculty candidate seminars were found on public departmental websites. Pub records were acquired from Google Scholar or Pubmed, and K99's were found on NIH Reporter. I did my best to summarize research topics by reading the abstracts of a candidate's recent papers.


https://twitter.com/JSheltzer/status/1287084519846293505
 One of the giants in the field of cancer genetics is Peter Duesberg, a member of the National Academy and a professor at UC Berkeley. He did pioneering work on SRC and retroviral oncogenes in the 70’s. Now he works on aneuploidy and cancer. BUT…
One of the giants in the field of cancer genetics is Peter Duesberg, a member of the National Academy and a professor at UC Berkeley. He did pioneering work on SRC and retroviral oncogenes in the 70’s. Now he works on aneuploidy and cancer. BUT… 
 Several people responded to this tweet by claiming you couldn't be nominated posthumously. This is also incorrect. Many examples - pathologist Katsusaburo Yamagiwa was nominated 6 years posthumously.
Several people responded to this tweet by claiming you couldn't be nominated posthumously. This is also incorrect. Many examples - pathologist Katsusaburo Yamagiwa was nominated 6 years posthumously.