
Cancer genomics, epigenomics and evolutionary dynamics (Weill Cornell/New York Genome Center). Hemato-oncologist (NYP). Discl.: https://t.co/tAu87FMVW3
How to get URL link on X (Twitter) App



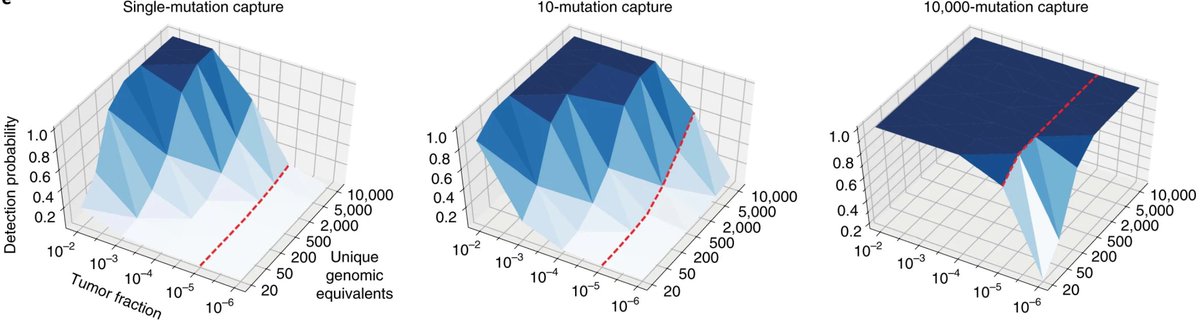


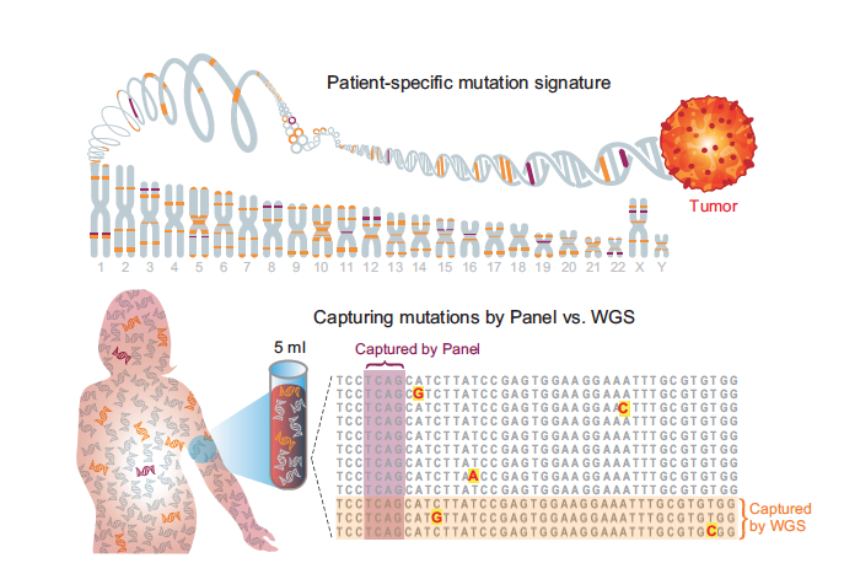
 Therapy monitoring is a central pillar of modern medicine. And yet, in many areas of oncology, we lack sensitive monitoring tools for residual disease detection. ctDNA carries the potential to change this, but is scarce in low-burden disease.
Therapy monitoring is a central pillar of modern medicine. And yet, in many areas of oncology, we lack sensitive monitoring tools for residual disease detection. ctDNA carries the potential to change this, but is scarce in low-burden disease.