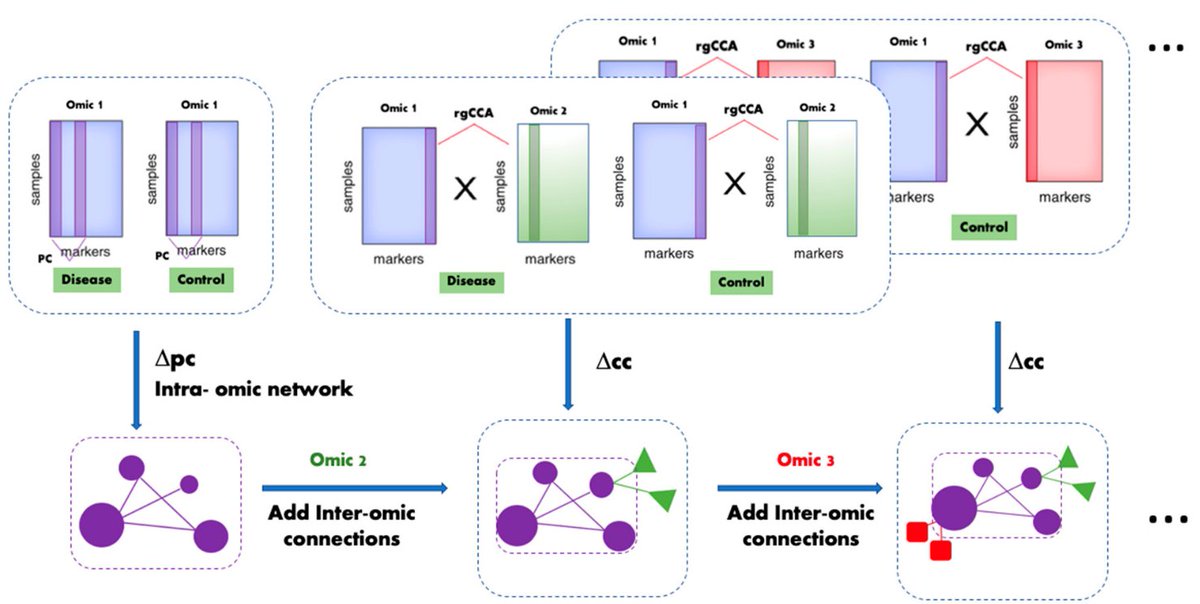
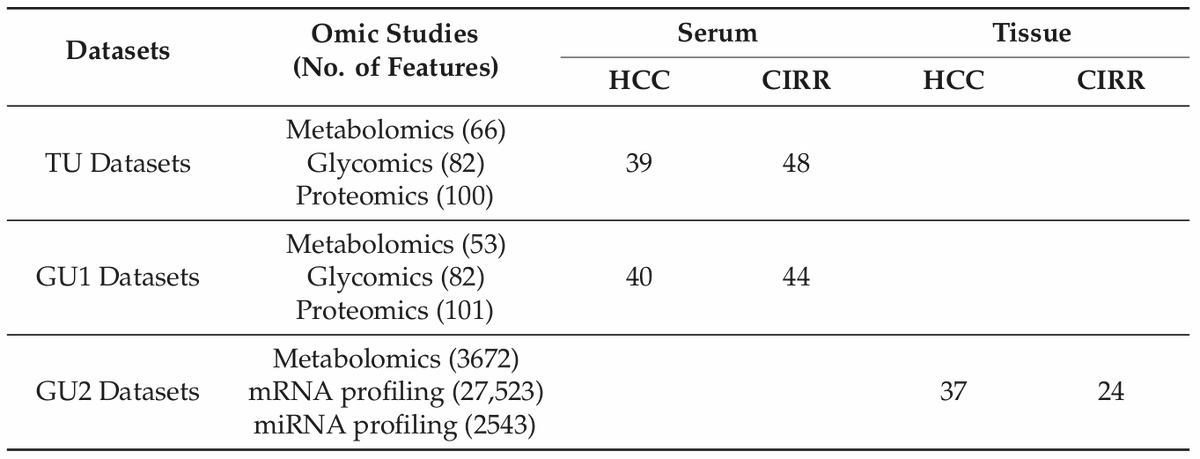
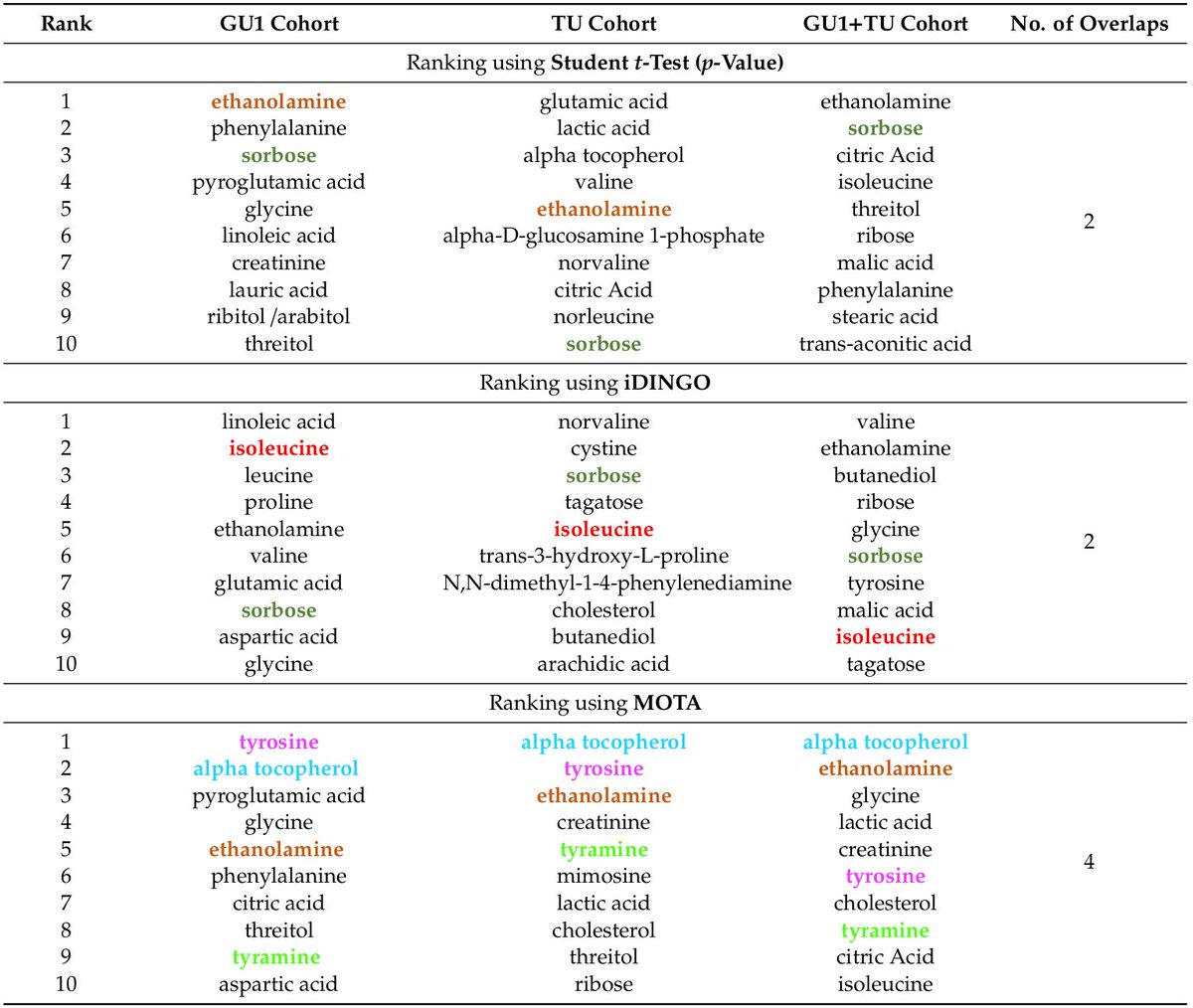
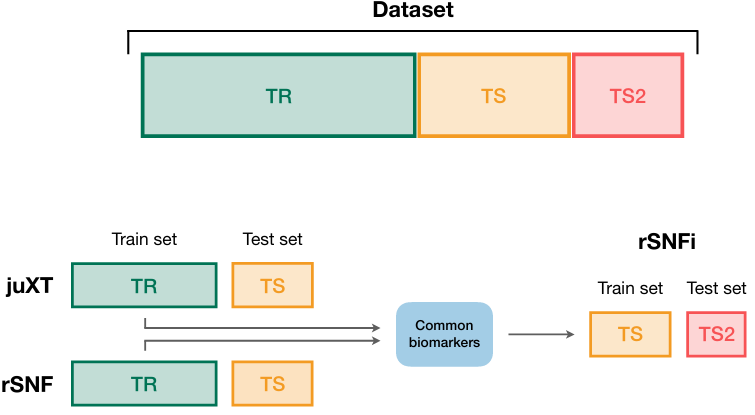
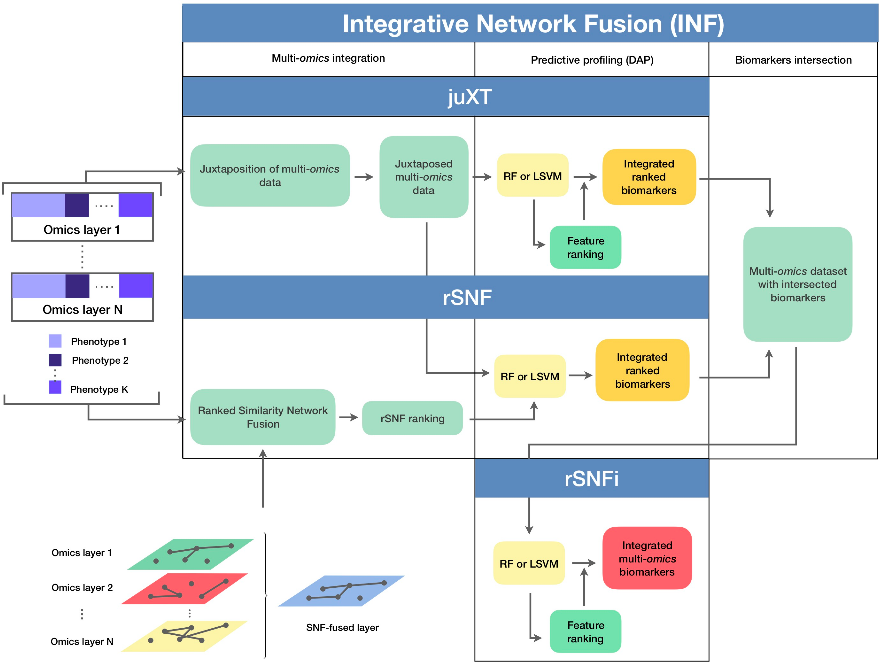
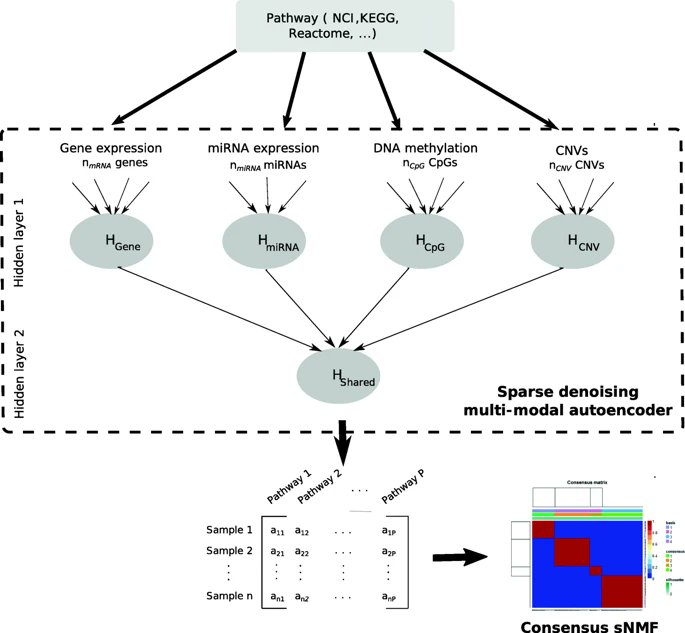
1) genes space → pathways space
2) for each pathway: collapse pathways from multiple omics to one per patient
3) sparse NMF biclustering
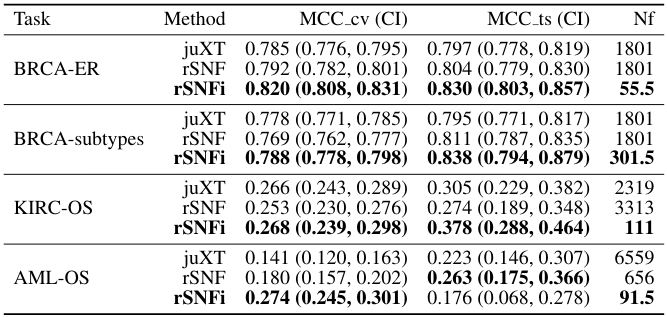
✓compared against SNF and iCluster
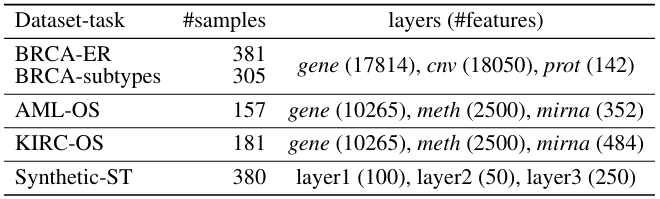
✓TCGA x 4
✓hyperparameters
✓source code
✓5-fold CV
#SundayMultiOmics
GitHub: github.com/AminaLEM/PathME
Figure © by authors, reused under CC-BY 4.0:
creativecommons.org/licenses/by/4.0
#omics #NeuralNetworks
- authors use sNMF consensus from 500 runs (cophenetic correlation + permutation testing to choose # of clusters)
- the autoencoders are denoising
- worth praise is the effort into interpretability (of both features/omics & clinical associations) - see supplement!
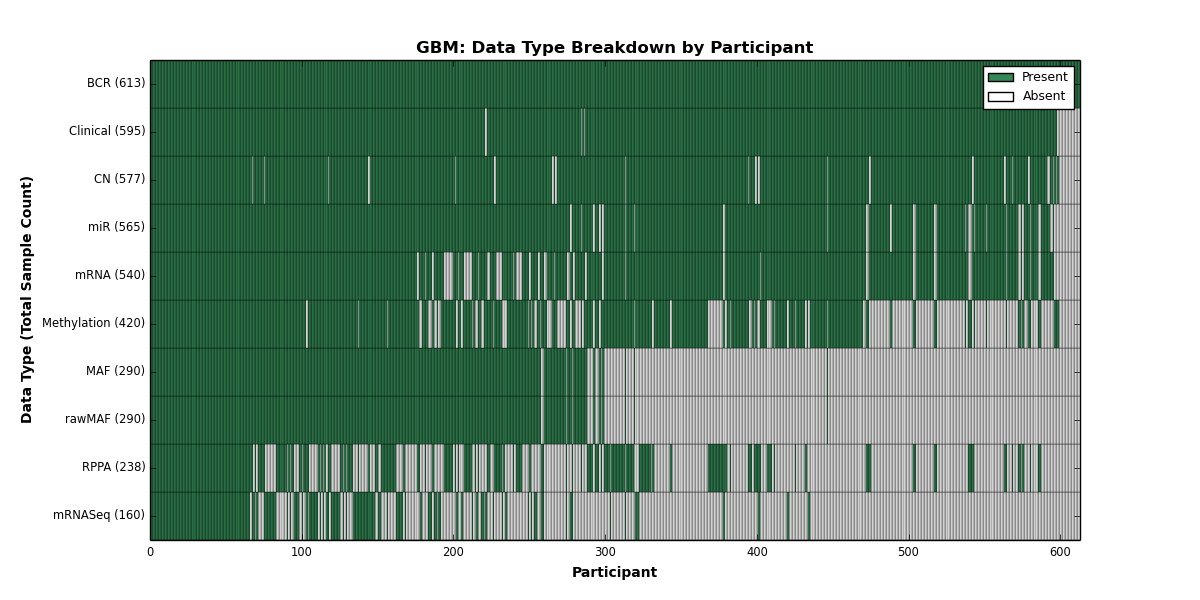
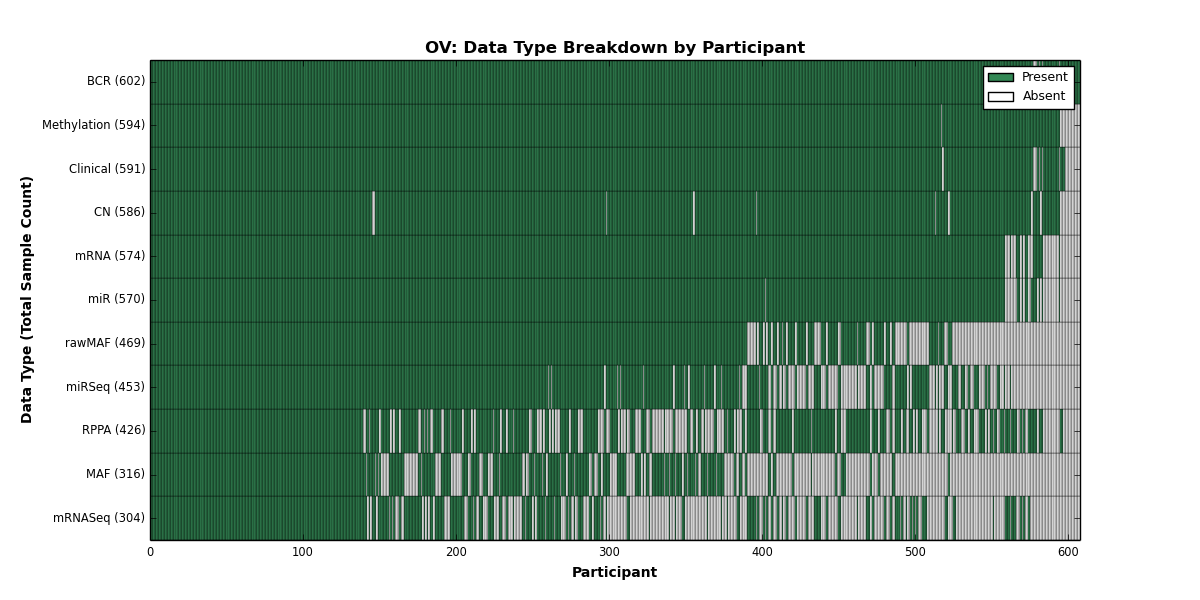
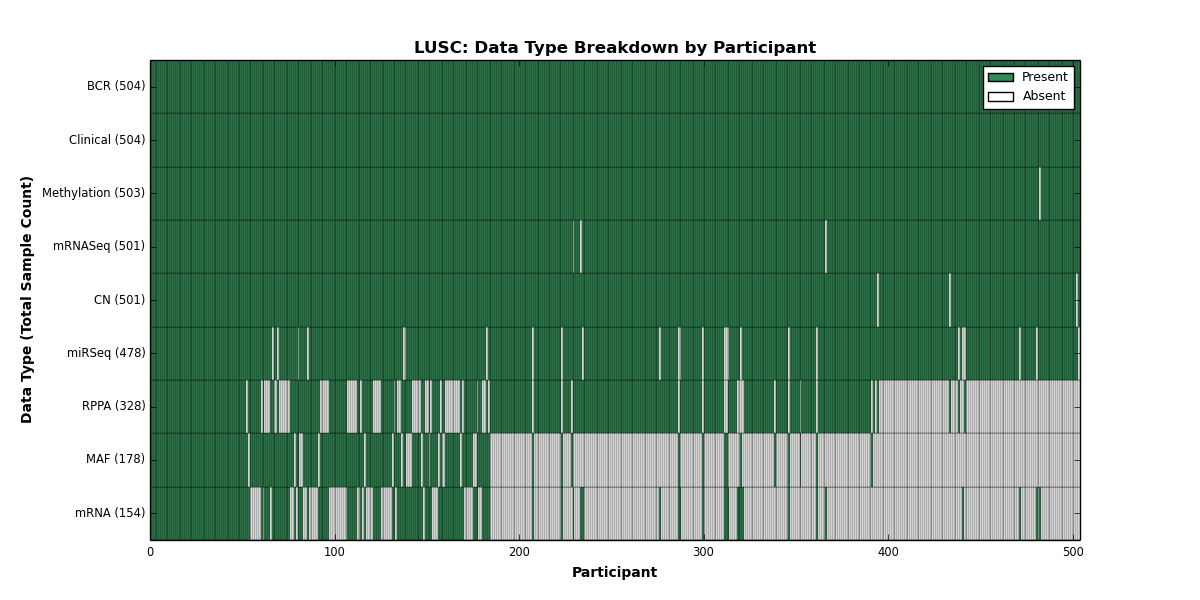
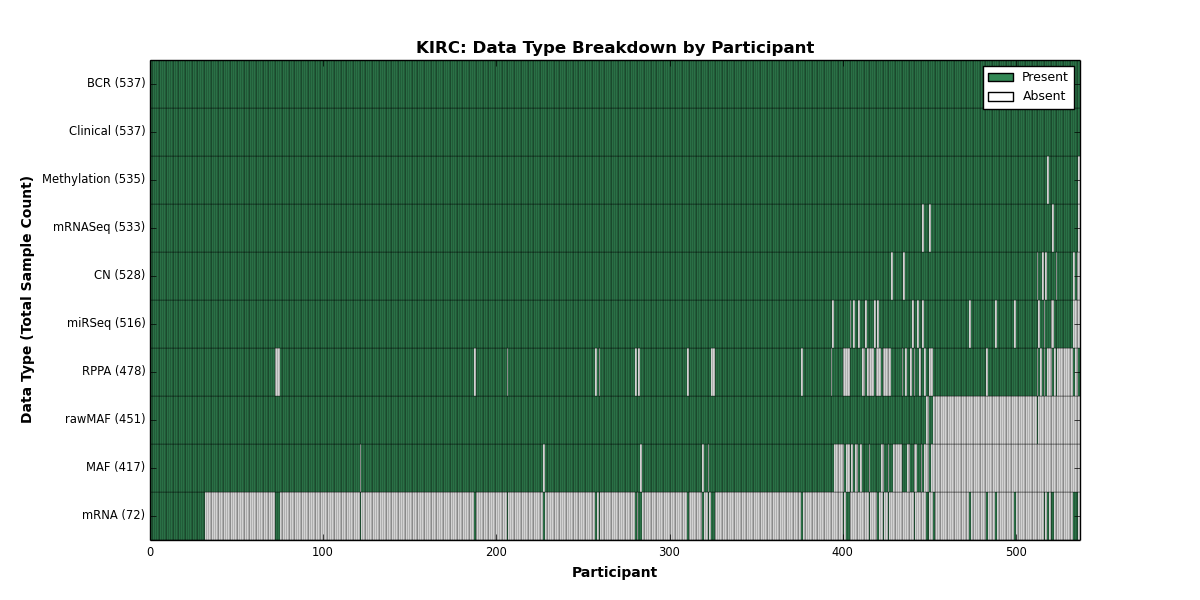
- golubEsets covariates are "of provenance unknown to the maintainer" (sic)
- only 24% of genes (1729) were actually mapped to pathways. Probably NCI Pathways is not the best idea...