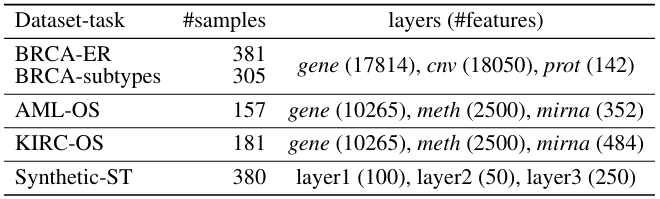
✓ 3 TCGA cancers & simulated data
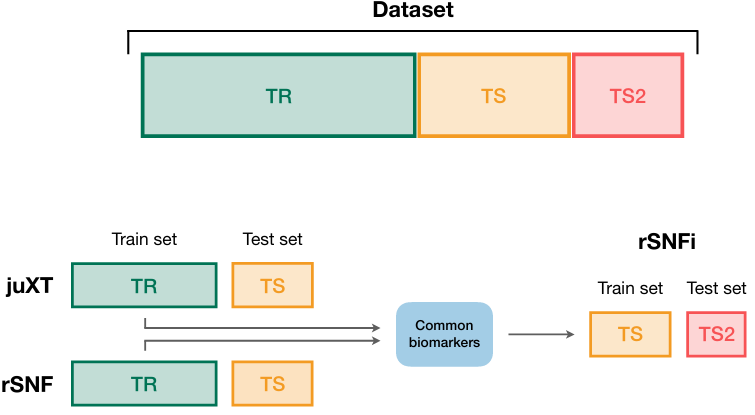
✓ cross-validation described in detail
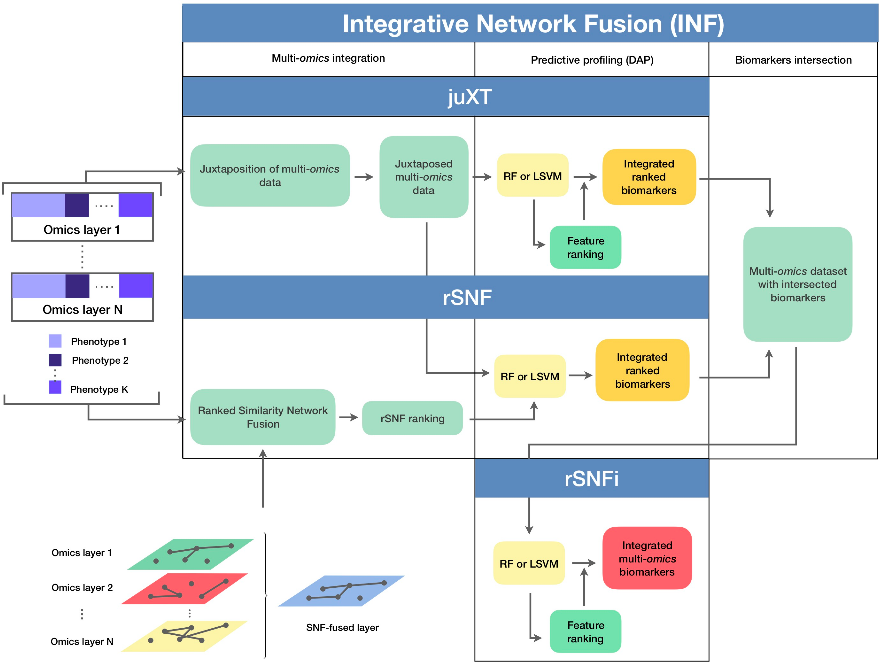
✓ flow diagram
✓ source code & data shared
✓ packages w/ version, cited
/n
- link: biorxiv.org/content/10.110…
- licence the above figures/tables: CC BY-NC-ND 4.0
- an earlier version of INF was previously presented in 2018: doi.org/10.1186/s13062…
- this is the first tweet in #SundayMultiOmics series
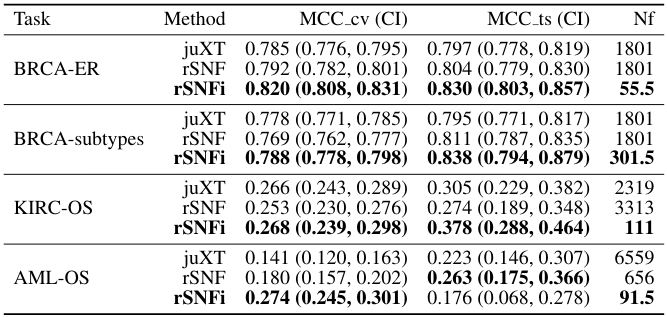
- breast invasive carcinoma: ER status, subtypes
- acute myeloid leukemia: overall survival
- renal clear cell carcinoma: overall survival
Comment: BRCA subtypes used (PAM50) are defined by transcriptomic signatures; as such the multi-omic methods...
- breast invasive carcinoma: #transcriptomics (mRNA), #proteomics (I guess RPPA), genomic data (copy number variation, #CNV)
- AML & KIRC: transcriptomics: mRNA & miRNA + #methylation
Note: an explanation of why those particular cancers/omics were chosen would be nice!
It seems that the accelerated version intentionally "leaks" data in the CV step...
/n







