Precision therapies for #cancer are underutilized for a variety of reasons: lack of sensitive diagnostic tools, lack of clinician awareness, et cetera.
This is changing one step at a time.
(A Thread)
This is changing one step at a time.
(A Thread)
Conceptually, precision drugs exploit genetic defects in a patient's tumor. Using comprehensive next-gen #sequencing (NGS), clinicians can surface mutations that indicate the use of a precision drug.
If we don't catch mutations, we underutilize therapies.
If we don't catch mutations, we underutilize therapies.
One notoriously difficult variant class is an #NTRK Fusion, which is common in certain pediatric cancers and sometimes present in common adult cancers.
While precision therapies targeting NTRK-fusions exist, only recently have we improved our ability to detect them in tumors.
While precision therapies targeting NTRK-fusions exist, only recently have we improved our ability to detect them in tumors.
NTRK is an umbrella term for three genes (NTRK1, 2, and 3) responsible for the growth of nerve tissue. NTRK genes encode for transmembrane proteins (TRK, as shown below). TRK proteins 'catch' growth signals and relay that signal to the cell nucleus triggering cell growth! 

An NTRK-fusion happens when part of the NTRK gene collides with another gene. This makes a 'Frankenstein' (chimeric) protein. Functionally, this is like kicking in a light switch such that the light is always on.
In a cancer cell, this means the 'grow!' signal is always on.

In a cancer cell, this means the 'grow!' signal is always on.


Why are these hard to detect? Firstly, sequencing DNA isn't the right choice because the non-coding (intronic) regions of NTRK genes are highly repetitive, which biases #sequencing results.
A better choice is to sequence RNA, but this still leaves us with a big problem...
A better choice is to sequence RNA, but this still leaves us with a big problem...

Before sequencing, we need to attach molecules (primers) to the edges of an mRNA. It's easy to bind one to the NTRK-side of the mRNA, but unless we know the other side of the fusion a priori, it's hard to bind a primer.
This means the mRNA can't be read very well.

This means the mRNA can't be read very well.


Anchored Multiplex PCR is a method that uses universal primers that always bind to the unknown (?) half of the NTRK fusion product, vastly improving analytical sensitivity by eliminating false negatives. 

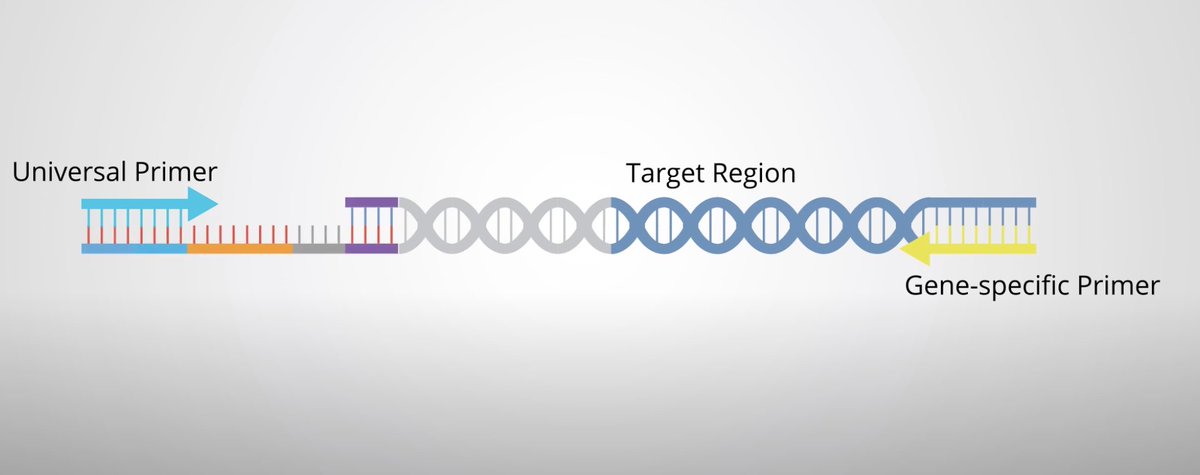
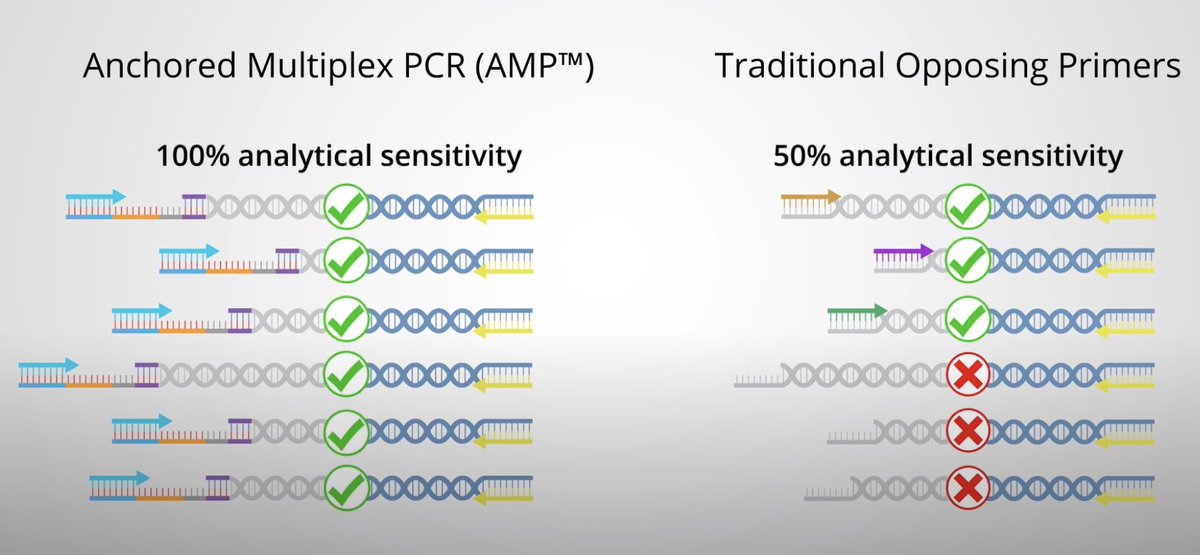
In summary, this is an example of a novel technique to more reliably call novel NTRK-fusion events in cancerous tissue, thereby increasing the utilization of TRK-inhibitors in those patients, ostensibly improving their outcomes.
• • •
Missing some Tweet in this thread? You can try to
force a refresh