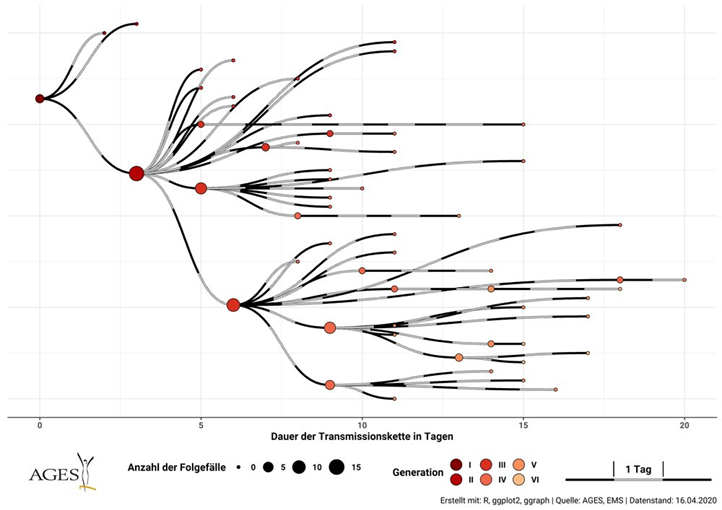
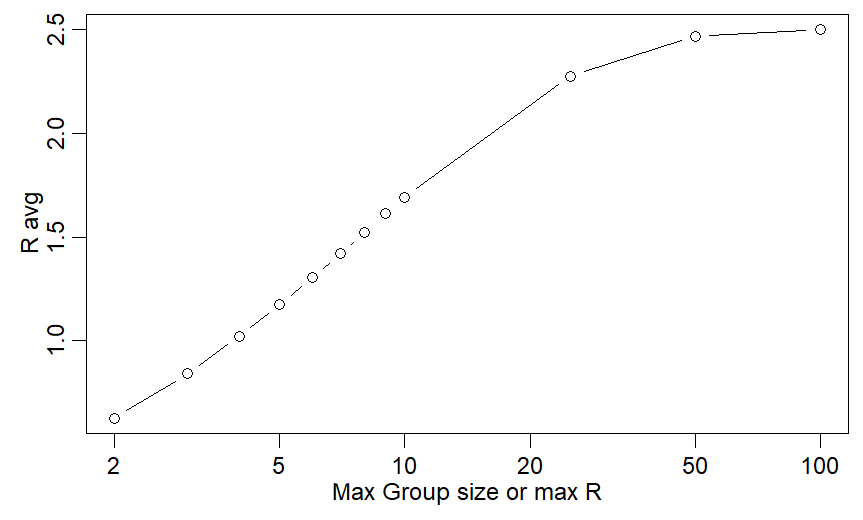It's now well known that SSEs play key role in #COVID19. Big Q: how do interventions reduce size of these events and how effective would they be?
Statsy/mathy thread.
sciencemag.org/news/2020/05/w…
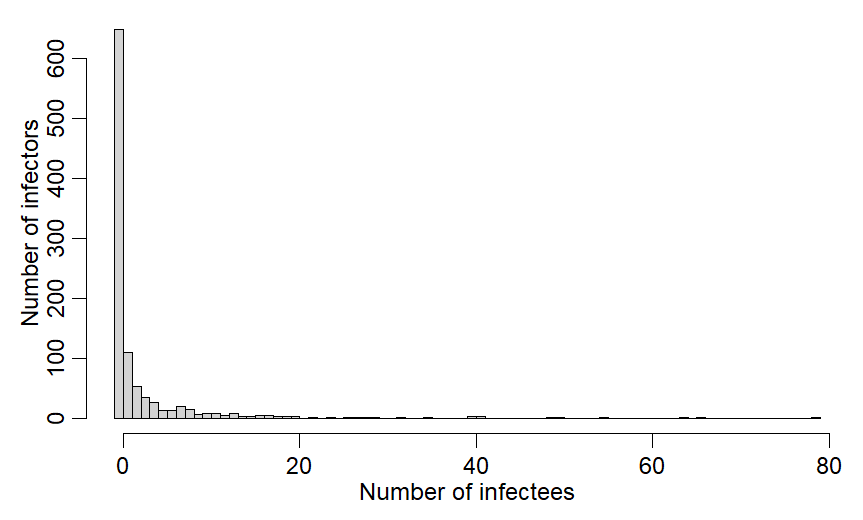
But the average is deceiving. Majority of cases infect no one; a few infect many!
x=rnbinom(1000,mu=R0v,size=0.16)
hist(x,breaks=-1:(max(x)),main="",xlab="Number of infectees",ylab="Number of infectors")
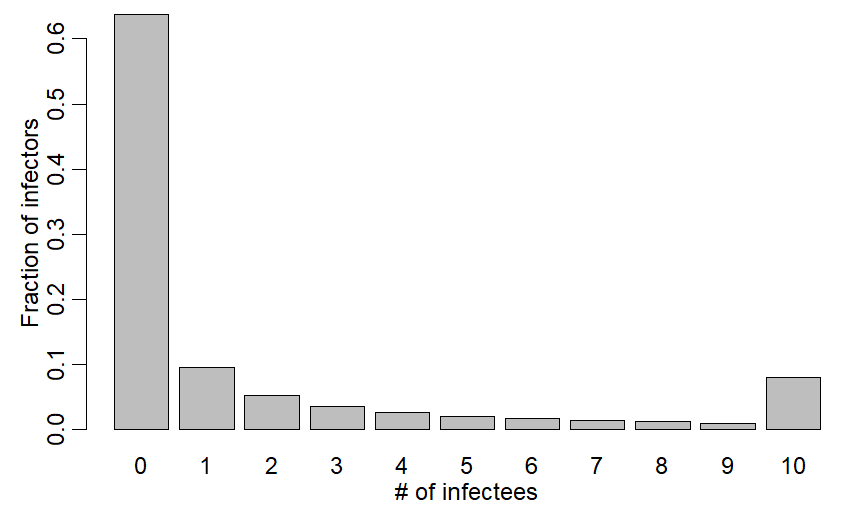
One thing many states & countries have done is cap # of people at events/gatherings. US used caps of 50,100. Childcare in CA now uses b/w 10-15. What effect would capped group sizes have on transmission/R?
Answer is a little more complicated than you think
xv=c(dnbinom(0:9,mu=2.5,size=0.16),1-pnbinom(9,mu=2.5,size=0.16))
barplot((xv),xlab="# of infectees",ylab="Fraction of infectors",names.arg=0:10)
Both model things a different way.
medrxiv.org/content/10.110…
arxiv.org/abs/2005.13689
arxiv.org/abs/2005.13689
(revised to use Ravg=2.5)
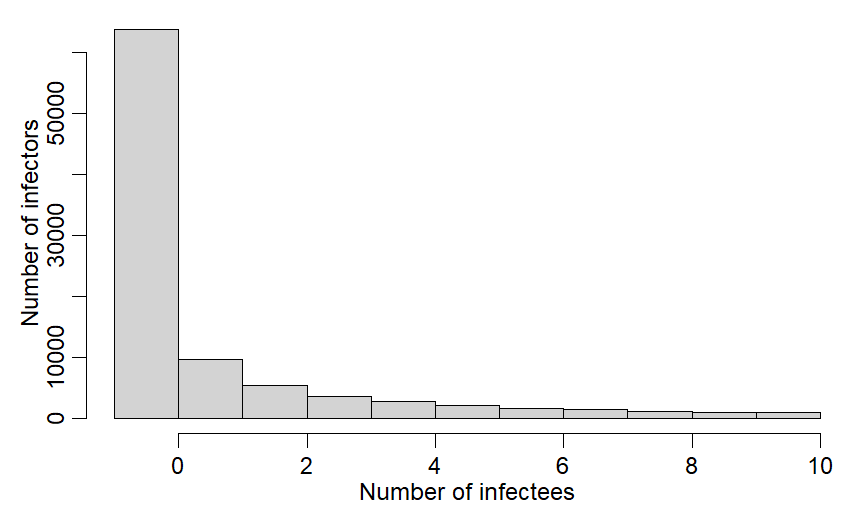
ss=100000
R0dist = rnbinom(ss, mu=2.5, size=0.16)
R0dist = R0dist[R0dist <=10]
sum(R0dist <=10)/ss
~8.5% of R vals are >10 & get excluded
If they are right, w/ Rmax=10, Ravg is just barely above 1 and cases are mostly stable - 5% growth/5d.
doi.org/10.1101/2020.0…
Thx to @BMAlthouse for email exchange (that I decided to share here after reading 2nd paper on this topic)
doi:10.1038/nature04153