As promised, here’s an overview of our study to find host factors required for SARS-CoV-2 infection in human cells. We use a genome-wide CRISPR screen to identify genes (& key mechanisms) that might be therapeutically targetable to prevent viral infection. 
Here’s a link to the manuscript (open access):
cell.com/cell/fulltext/…
cell.com/cell/fulltext/…
First & foremost: HUGE shout-out to first authors @ZDaniloski & #TristanJordan who worked incredibly hard to complete this study in a short time. Key ingredient was the great relationship btwn Zharko & Tristan: They made this happen. Full acknowledgements @ end of the 🧵.
Using our GeCKOv2 genome-wide CRISPR library (co-developed back in 2013 with @ophirshalem), we knocked out each of the ~20,000 genes in the human genome. 

We then challenged the genetically diverse pool of human lung cells with SARS-CoV-2. Key question: Which cells survive the virus and what CRISPR target enabled survival? 
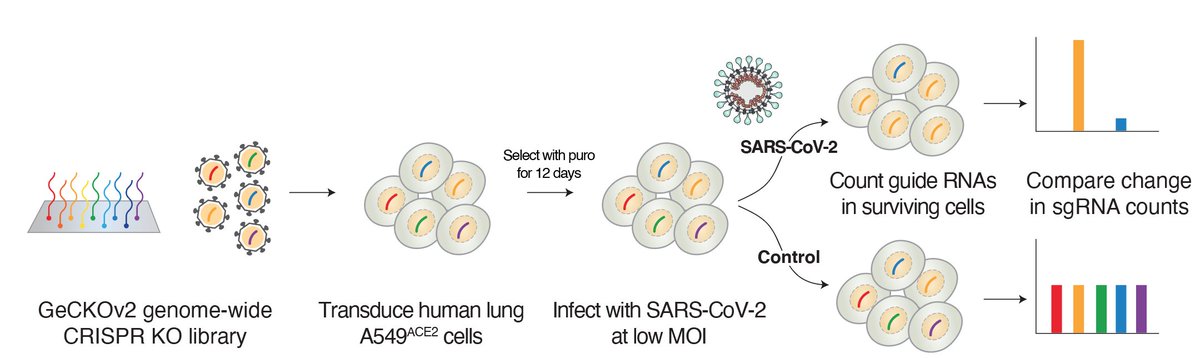
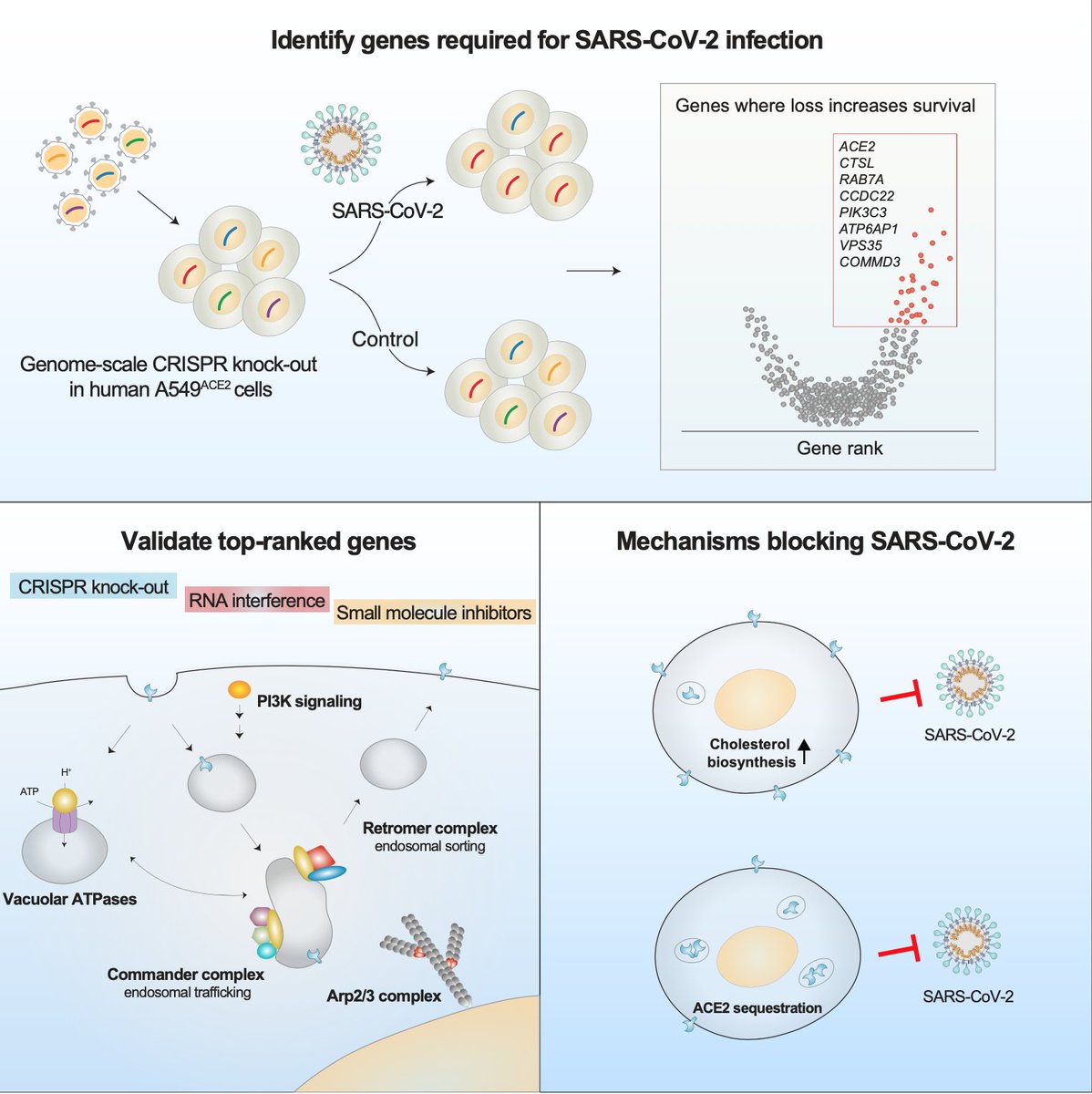
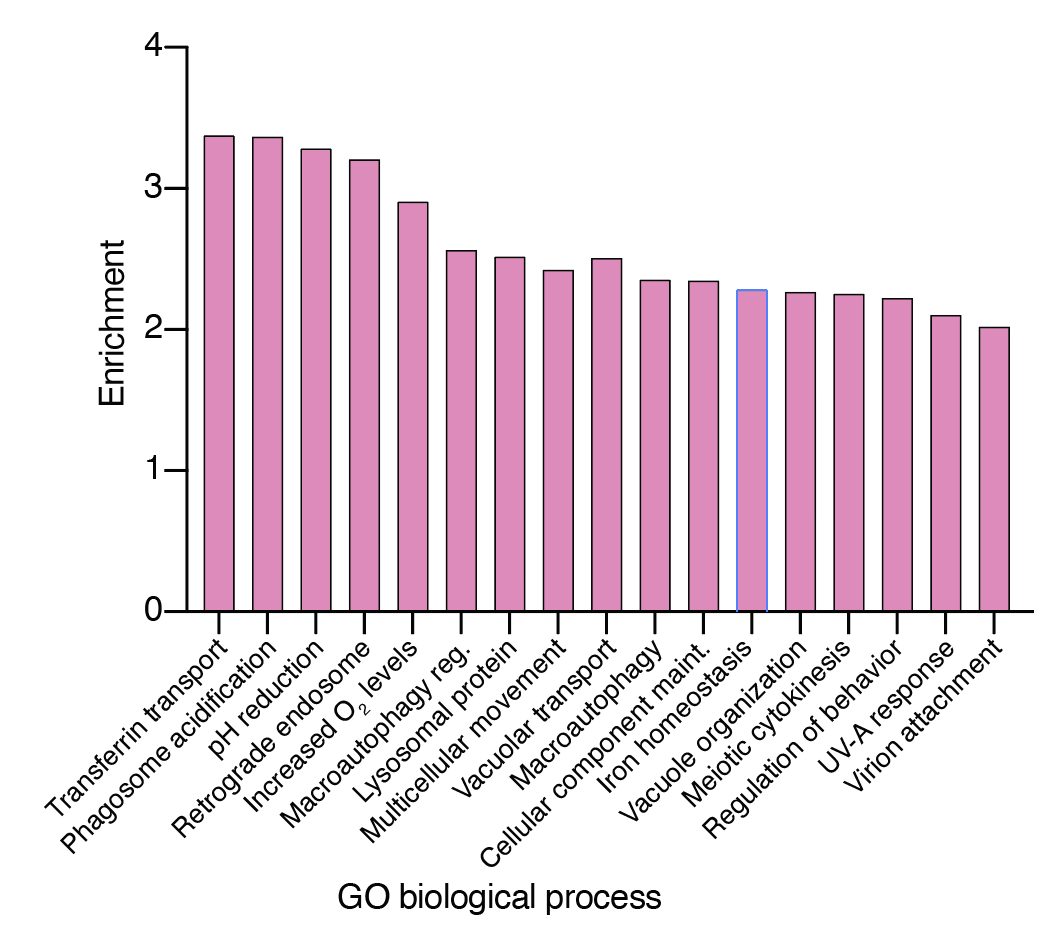
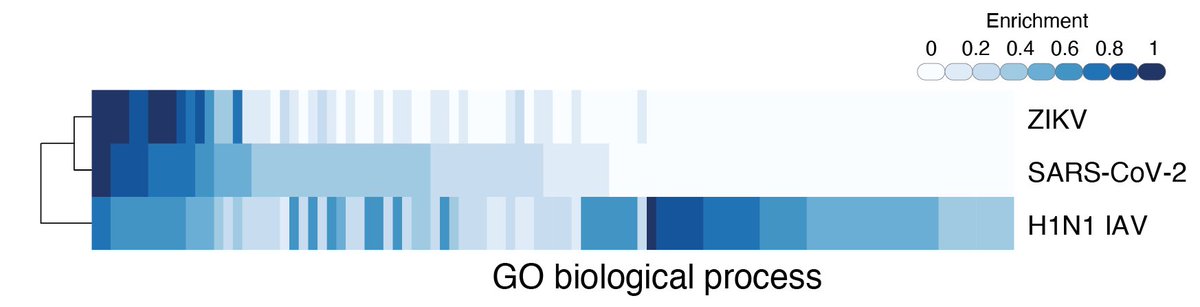
We found that top-ranked genes — genes whose loss increases resistance to viral infection — tended to cluster into specific gene families and pathways: 

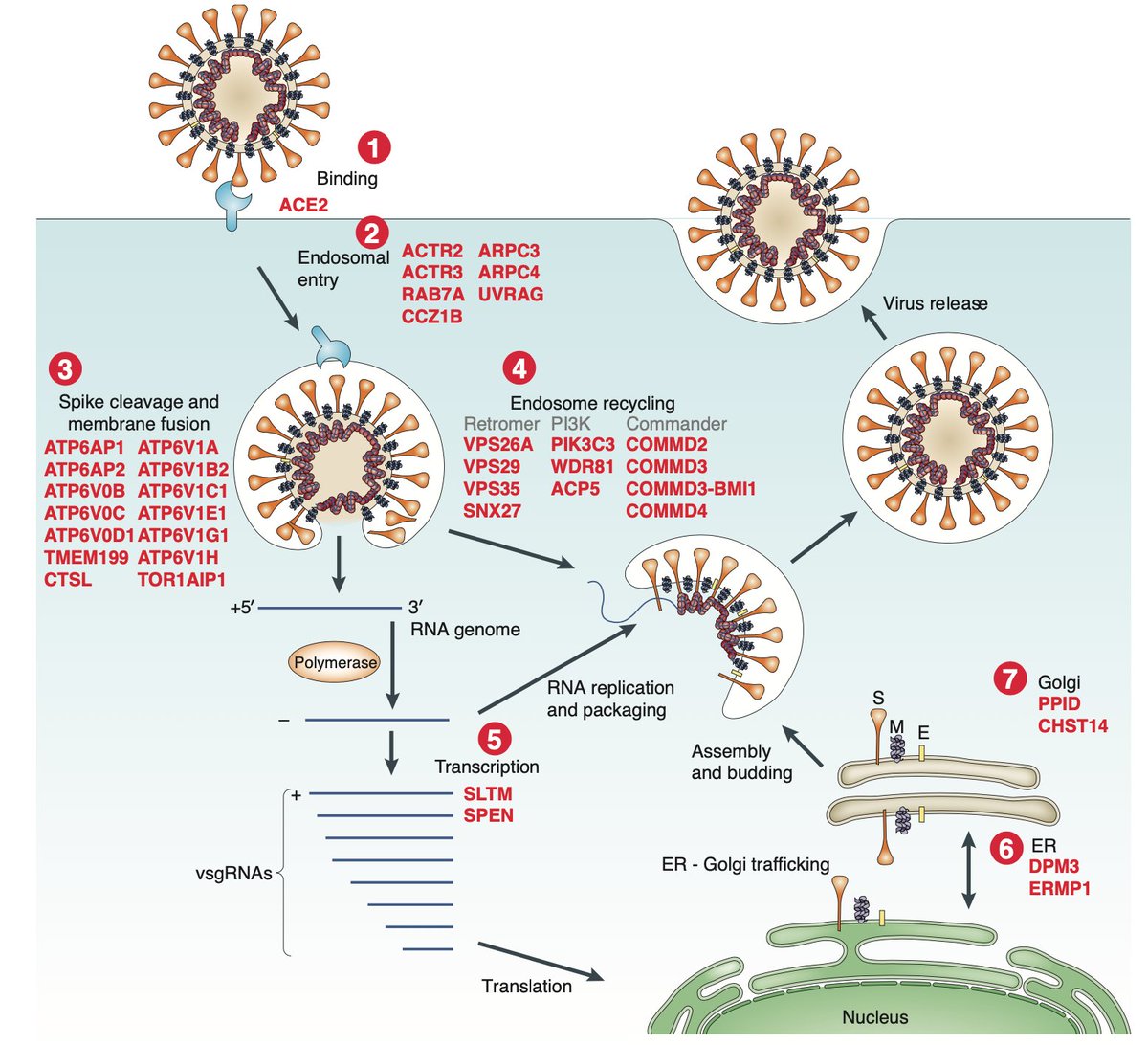
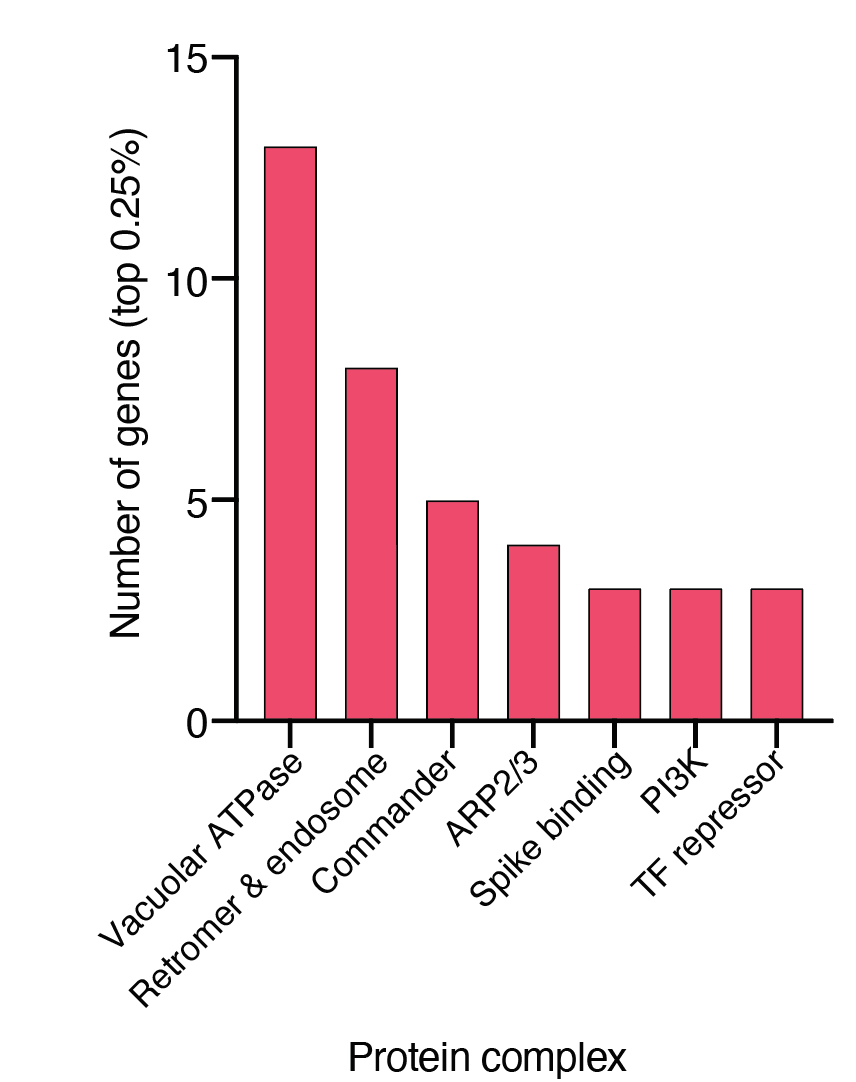
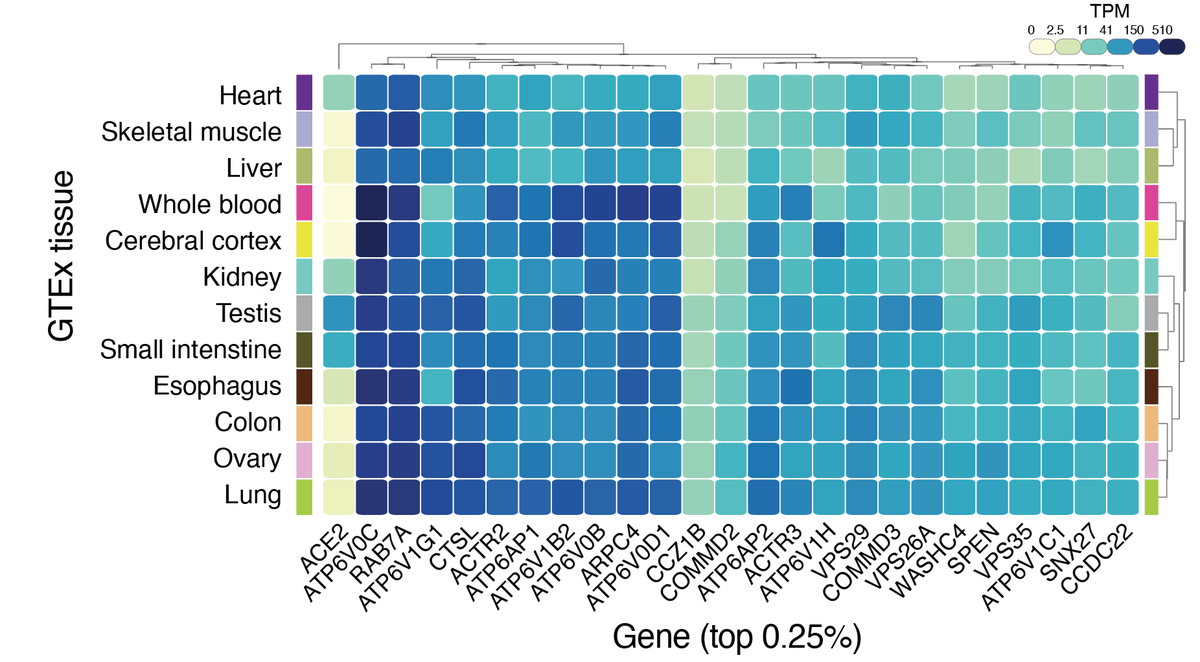
The majority of these top-ranked genes are broadly expressed across tissues and many are involved in intracellular transport & endosome function: 

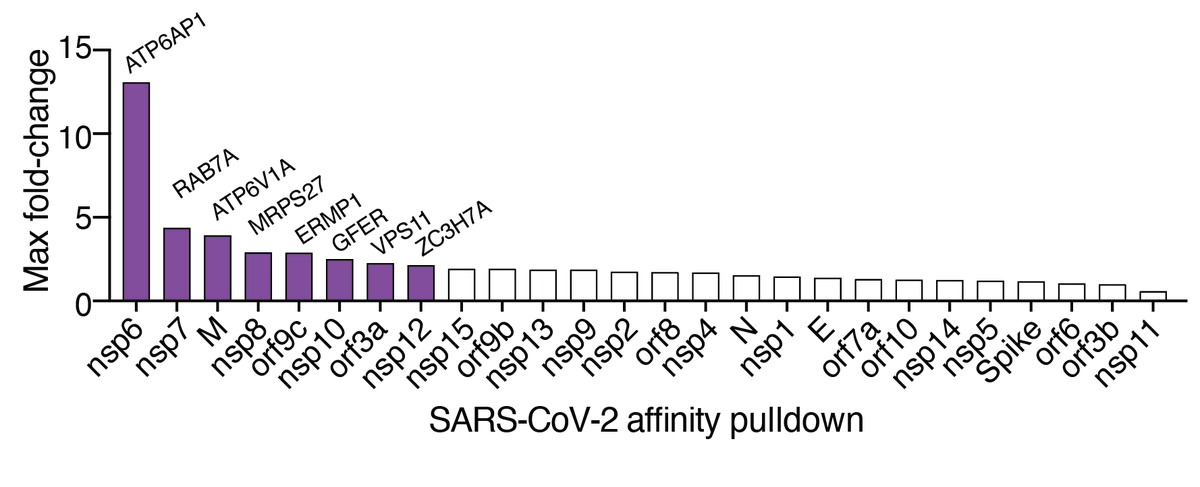
Using proteomics interaction data from @KroganLab, @gingraslab1, and #BrianRaught, we found that several of the top-ranked genes from our screen were previously shown to directly interact with SARS-CoV-2 viral proteins: 
We compared the results from the SARS-CoV-2 screen with prior CRISPR screens for Zika and pandemic (H1N1) flu from @yunli, #RudyJaenisch, @kennethbaillie, & #NirHacohen. We found shared gene hits from all 3 screens suggesting shared host mechanisms for resistance. 
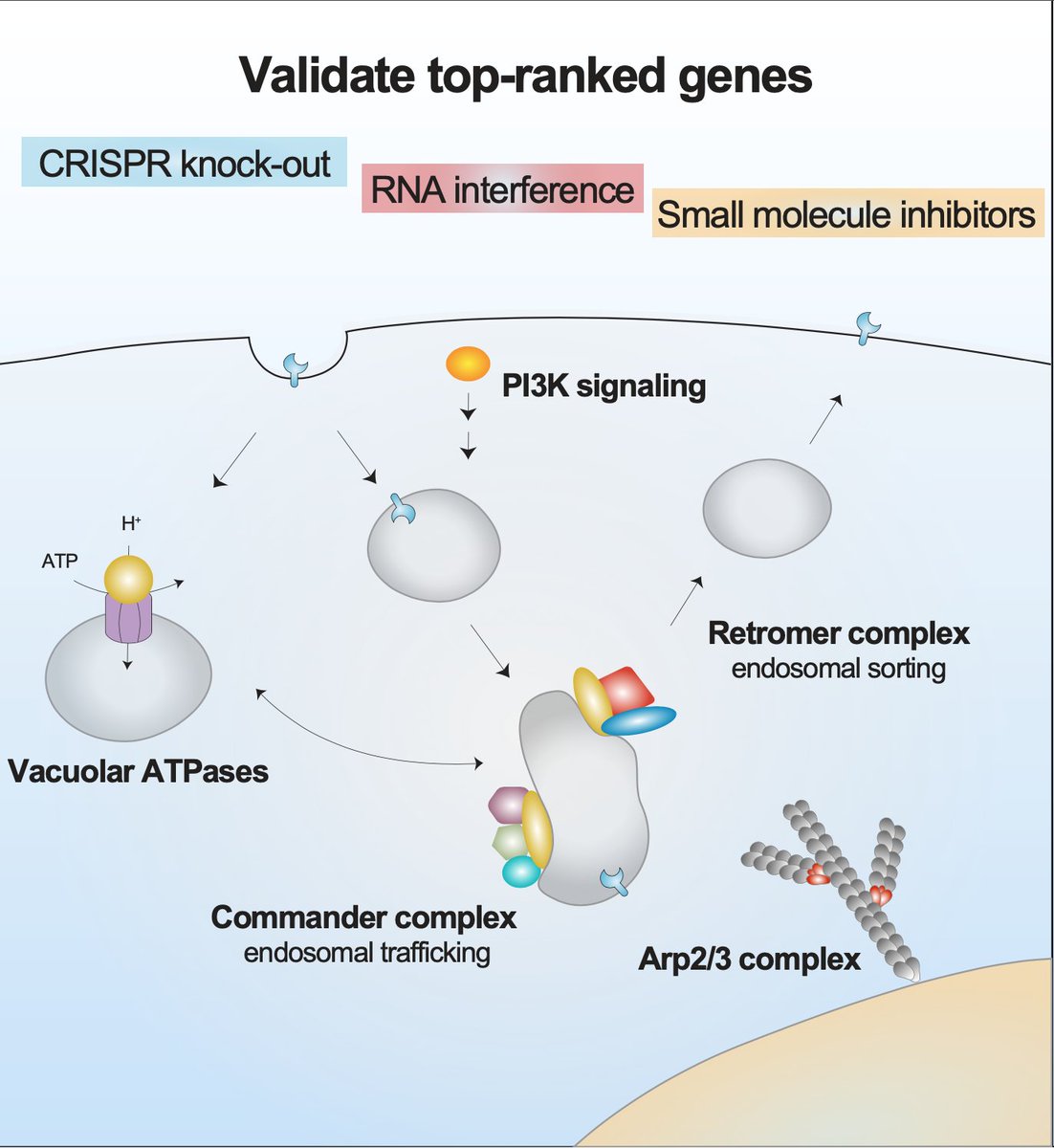
To validate our genome-wide screen and quantify the impact of each top-ranked gene on viral infection, we used several approaches, including gene knock-out (CRISPR), knock-down (RNAi), and small molecule inhibitors. 
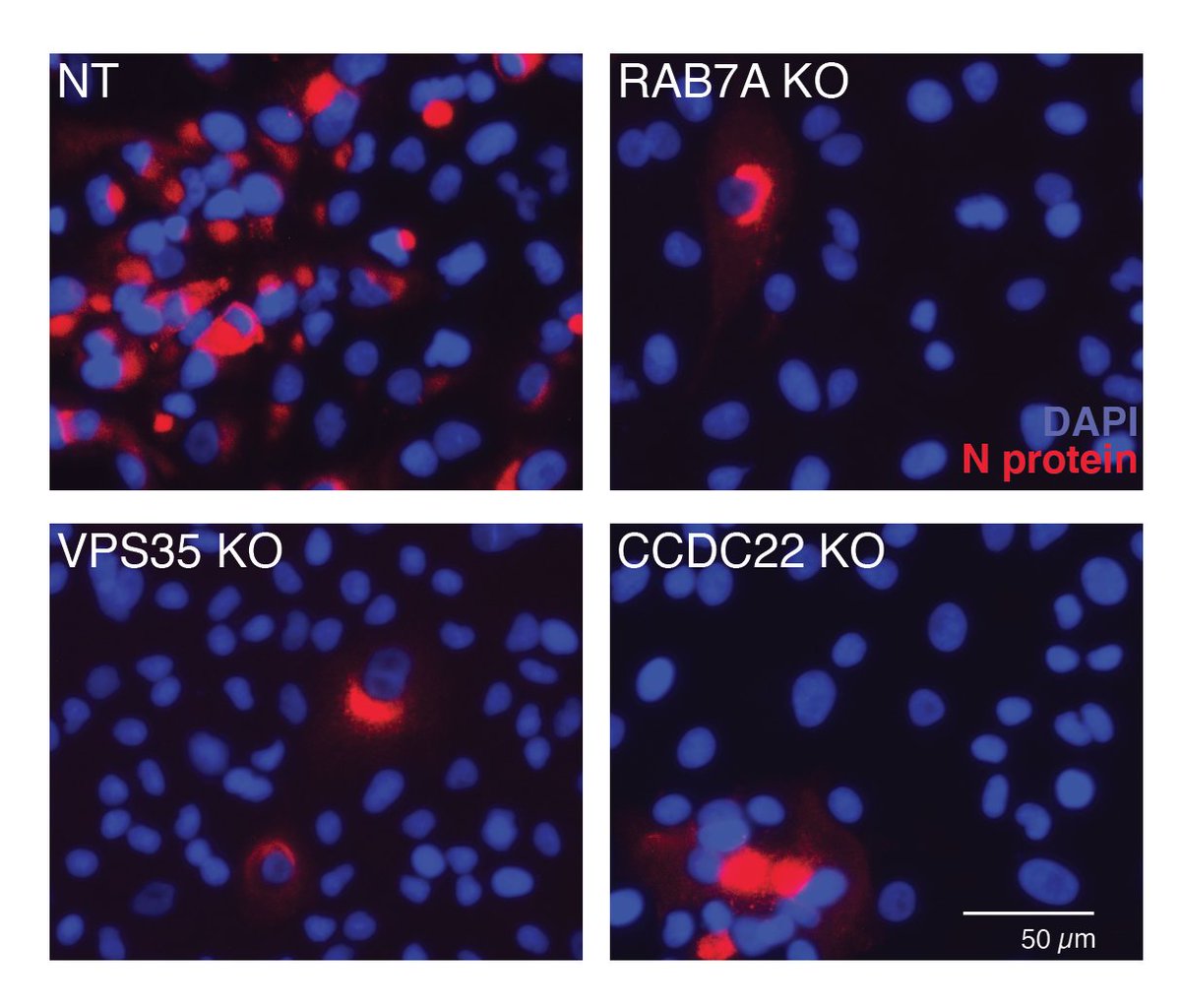
Here’s what some of the data looks like…. We counted the reduction in the number of cells infected by the virus after CRISPR knockout. 
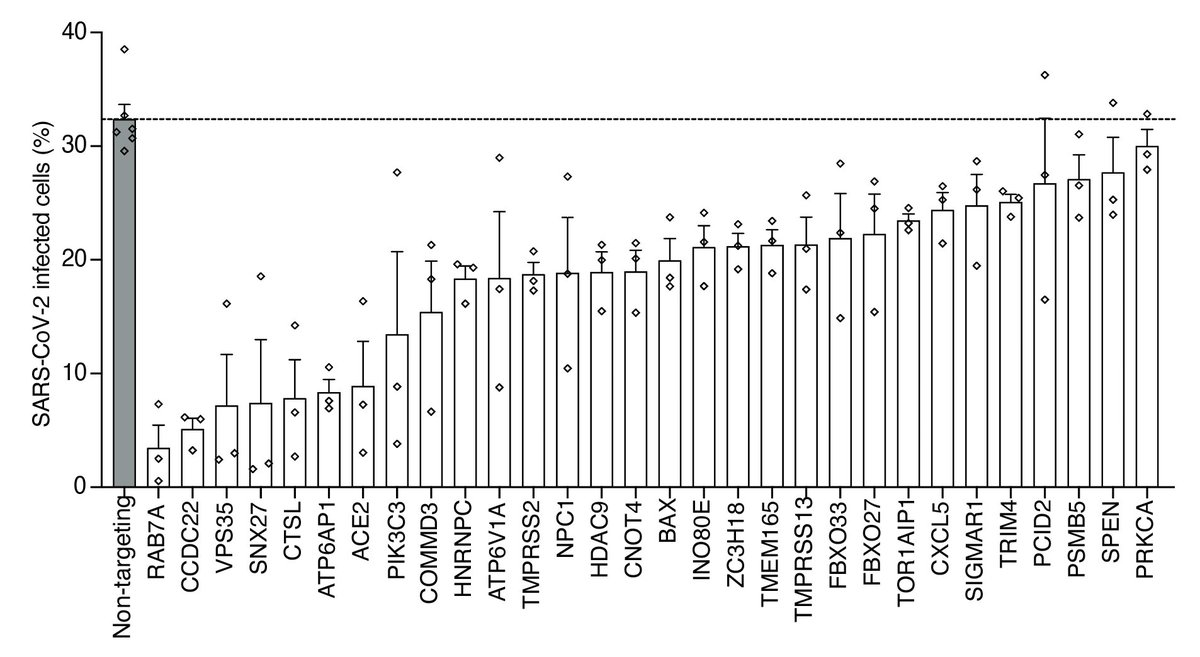
We sampled 30 genes from the top 200 genes from the genome-wide screen and basically all reduced infection — this was quite a surprise! 
Beyond understanding the biology of host factors, we wondered whether we could repurpose existing drugs that inhibit the same genes. Many of the top-ranked genes have commercially available inhibitors. Can we achieve the same results without using genome editing? 
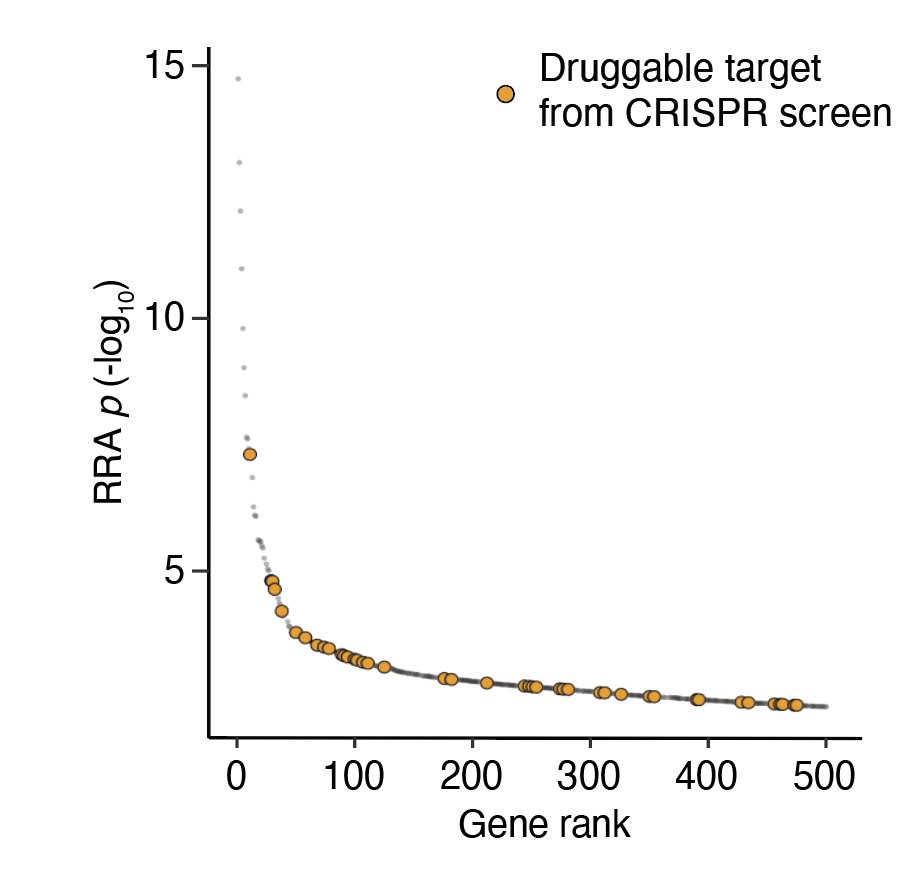
We took a handful of these and found that indeed many of them inhibit viral entry (and are not toxic to cells). Genetic mechanism leads the way to drug discovery! ⚙️💊🙂 
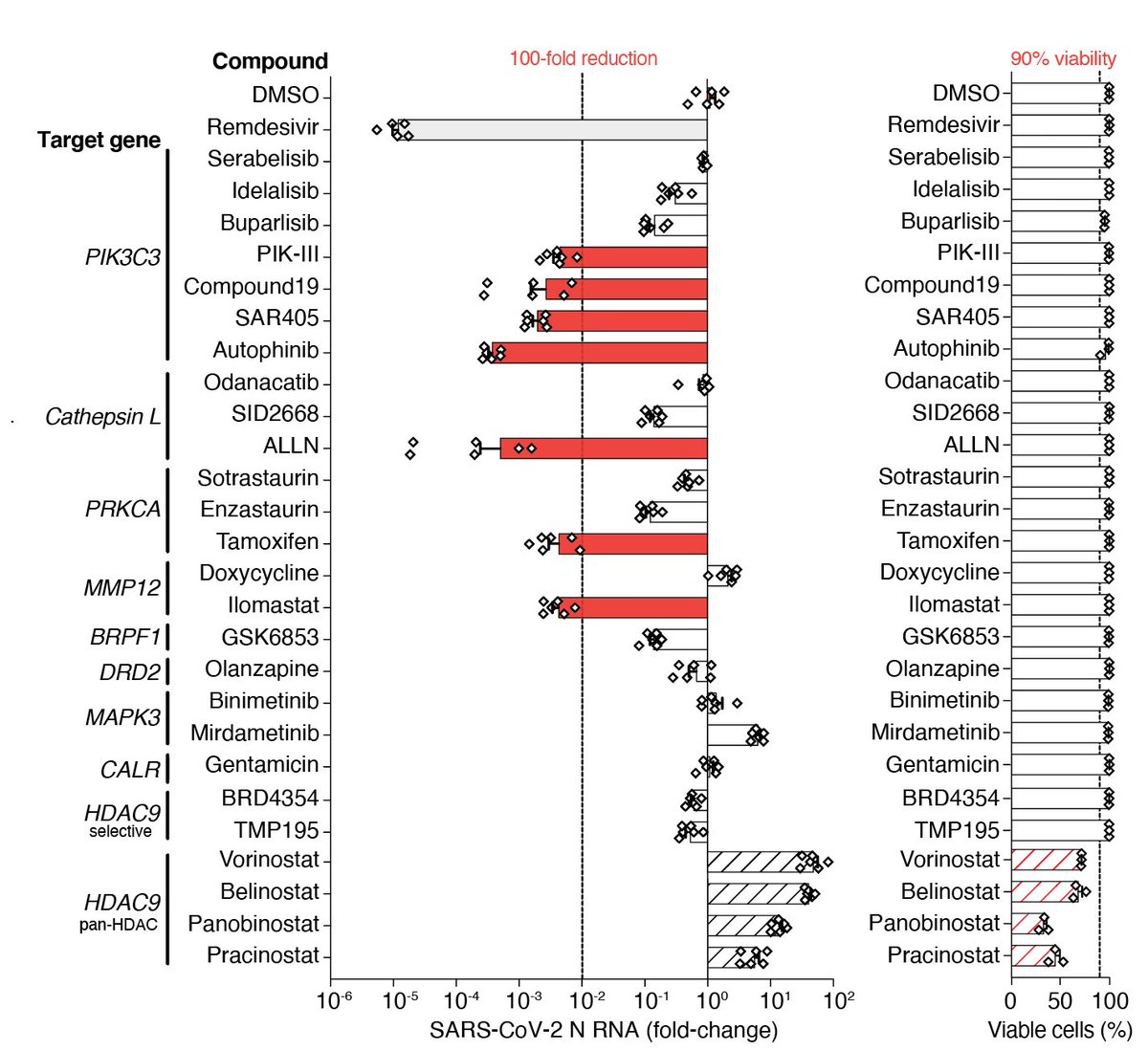
To get a better understanding of mechanisms that drive viral resistance, we decided to capitalize on ECCITE-seq (from our friends @NYGCtech) to pair CRISPR screens with single-cell RNA-sequencing. 
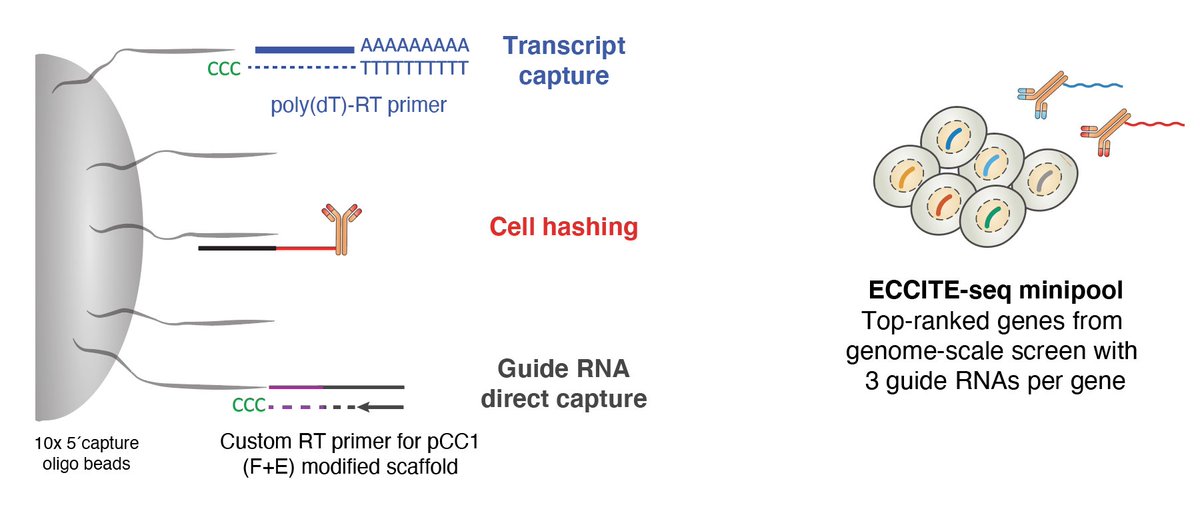
ENTER THE MATRIX! Looking at differential gene expression, we found that loss of some of the top-ranked genes created distinct changes in gene expression. Each column is a single-cell with a CRISPR targeting the gene shown on the top. 
But what does it mean?!? By looking for shared changes in gene expression, we saw that several CRISPR knockouts (KOs) modulate genes involved in cholesterol biosynthesis. 
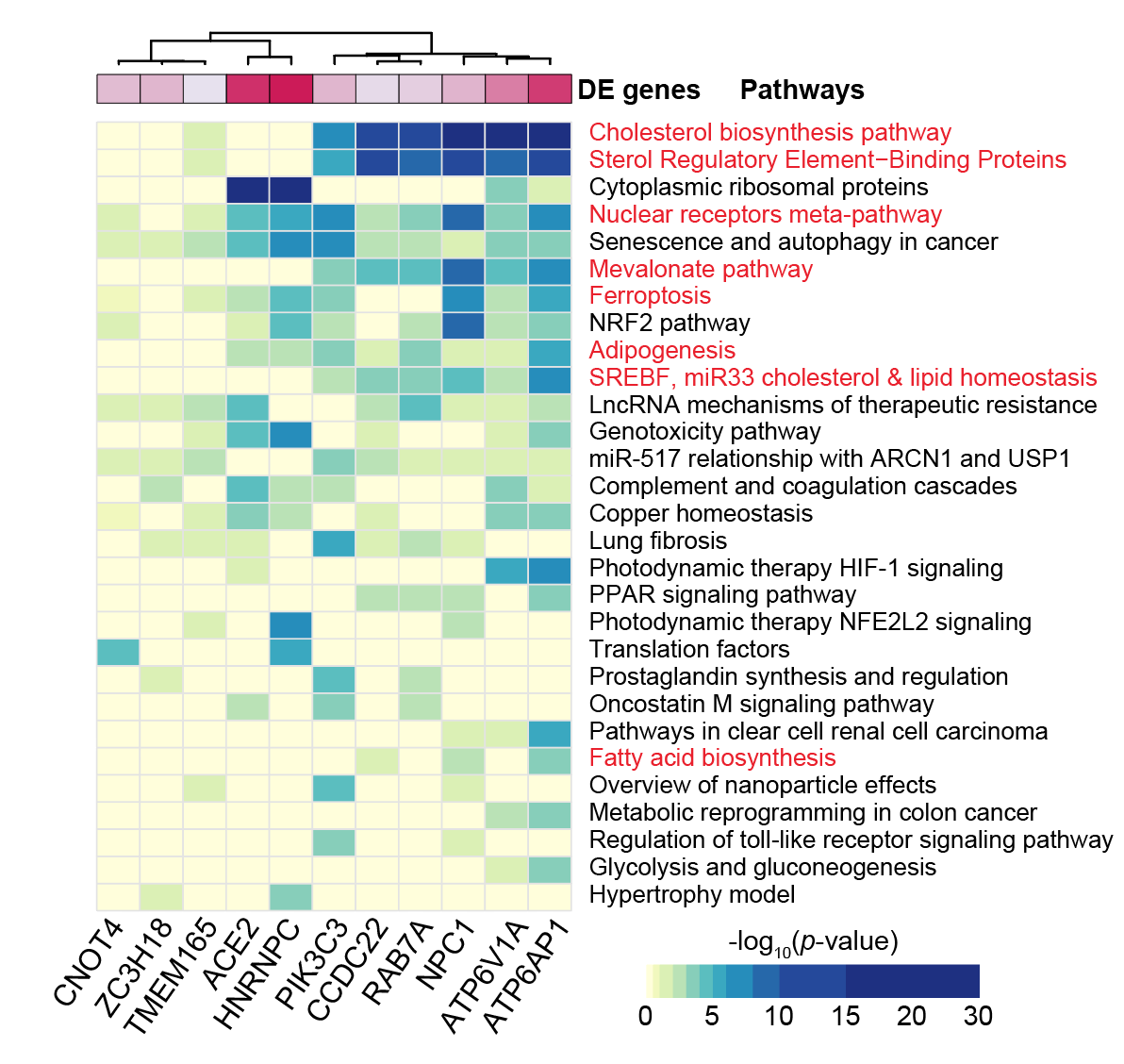
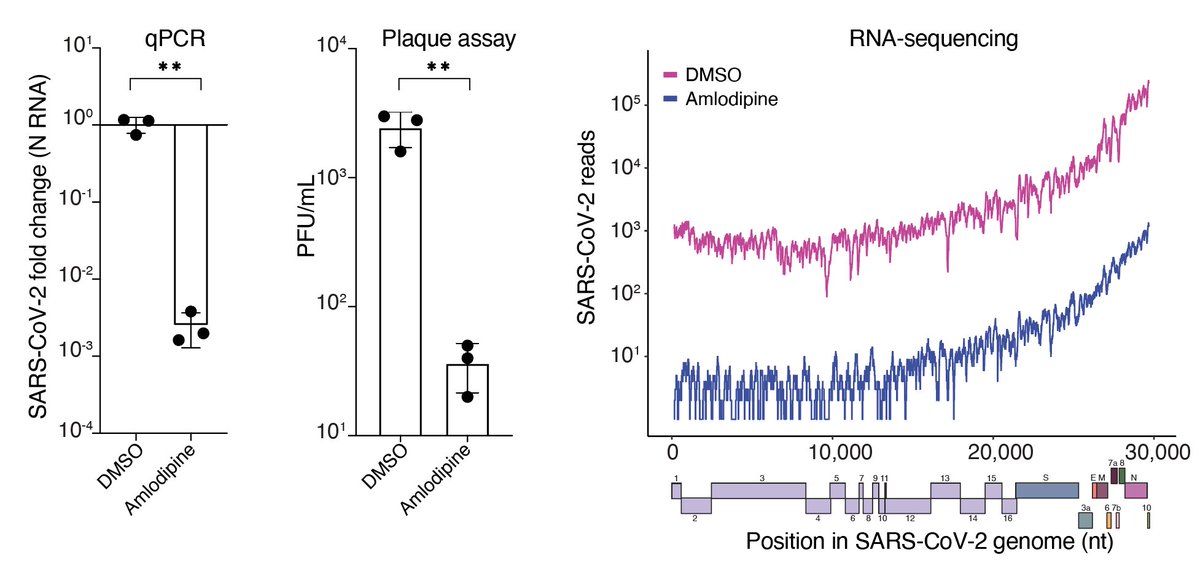
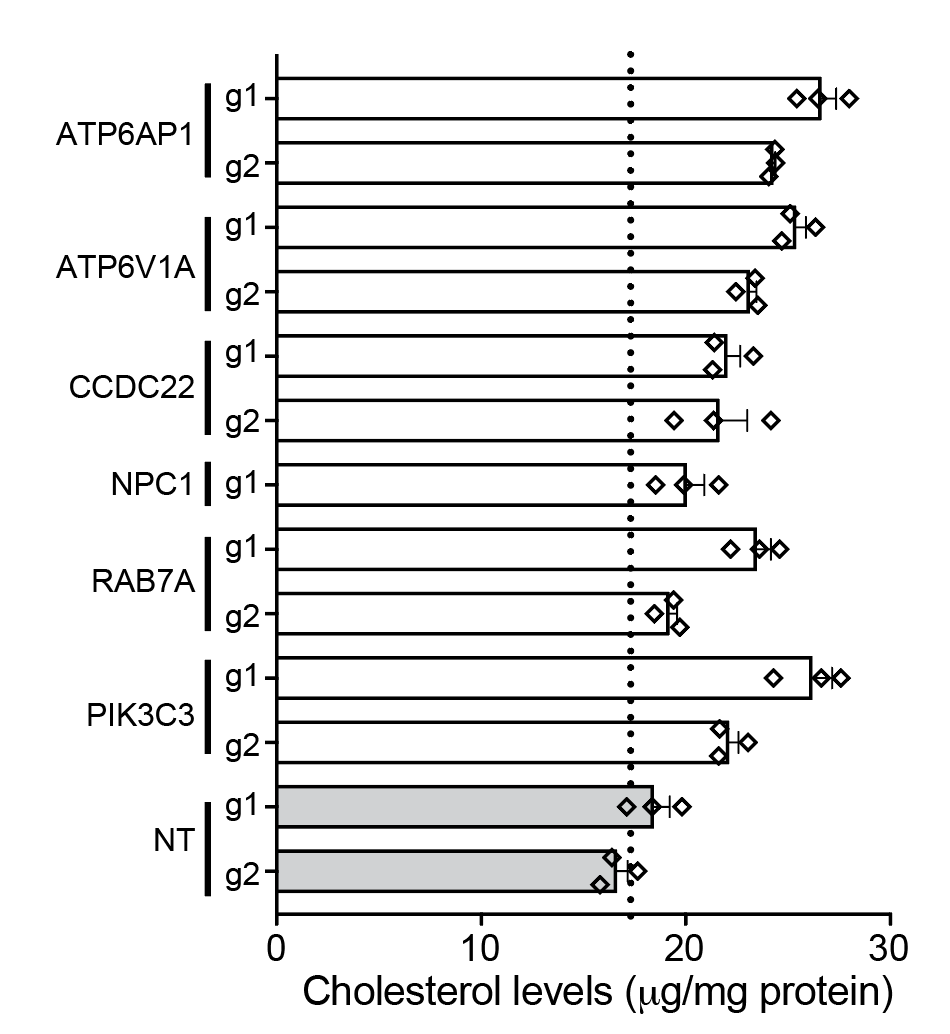
With this, we decided to measure cellular cholesterol after CRISPR KO. Guess what? All of those gene KOs increased cellular cholesterol…. ! 
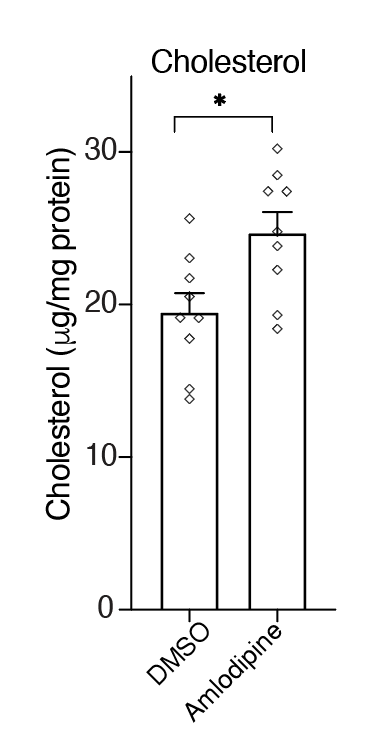
This suggested another therapeutic strategy: Can we mimic the increase in cholesterol without genome editing? Using the calcium channel blocker amlodipine, we found that we could increase cholesterol…. 
Next, given our previous work on Spike and ACE2 (bit.ly/preprint_05), we wondered whether some of the top-ranked genes might work via decreased ACE2 availability on the cell surface. 
We zeroed in on one gene, RAB7A, that appears to do just that. When cells lose RAB7A, the ACE2 on the cell membrane is reduced, which we can measure by flow cytometry. It seems that instead ACE2 just accumulates inside the cell. 


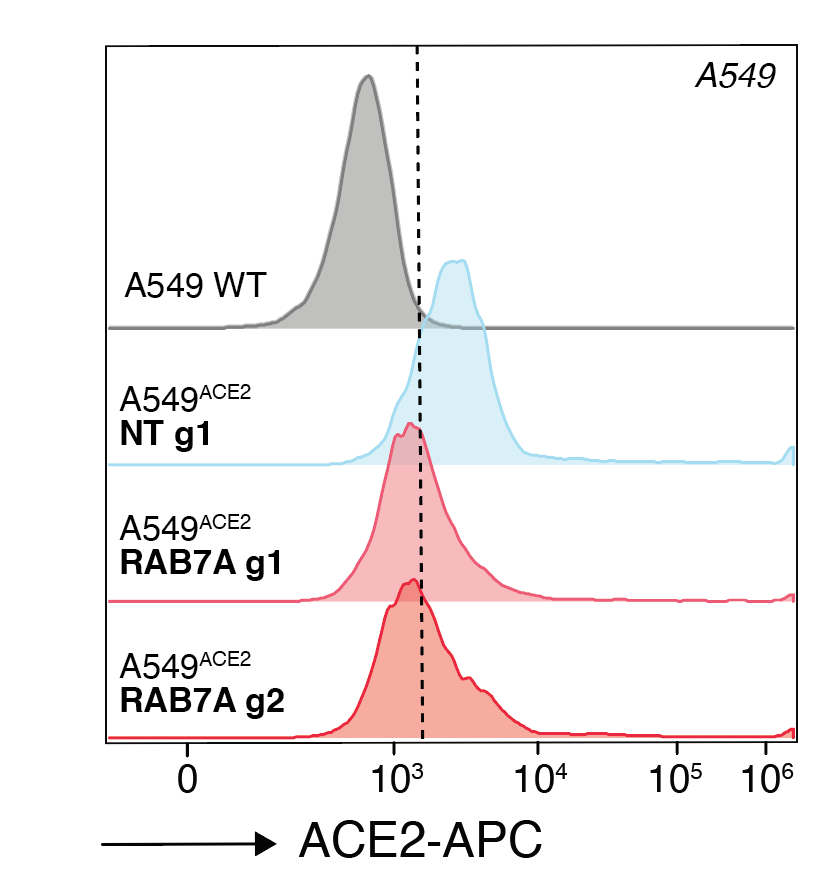
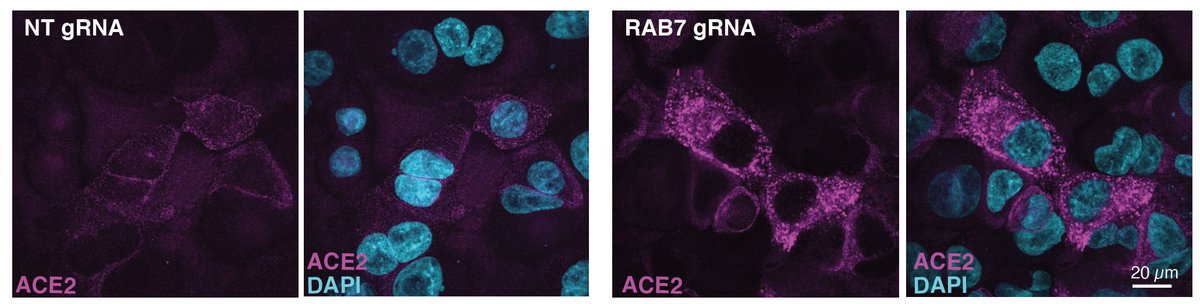
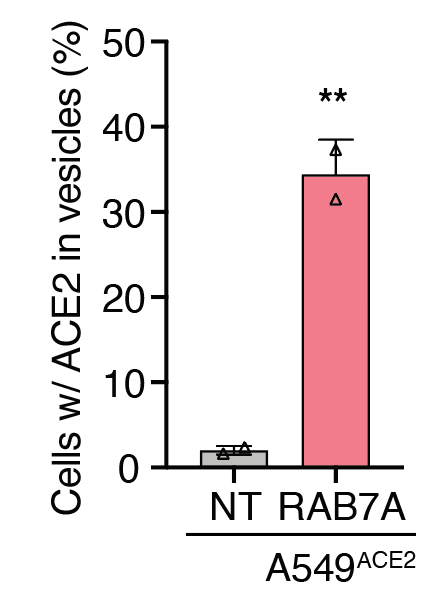
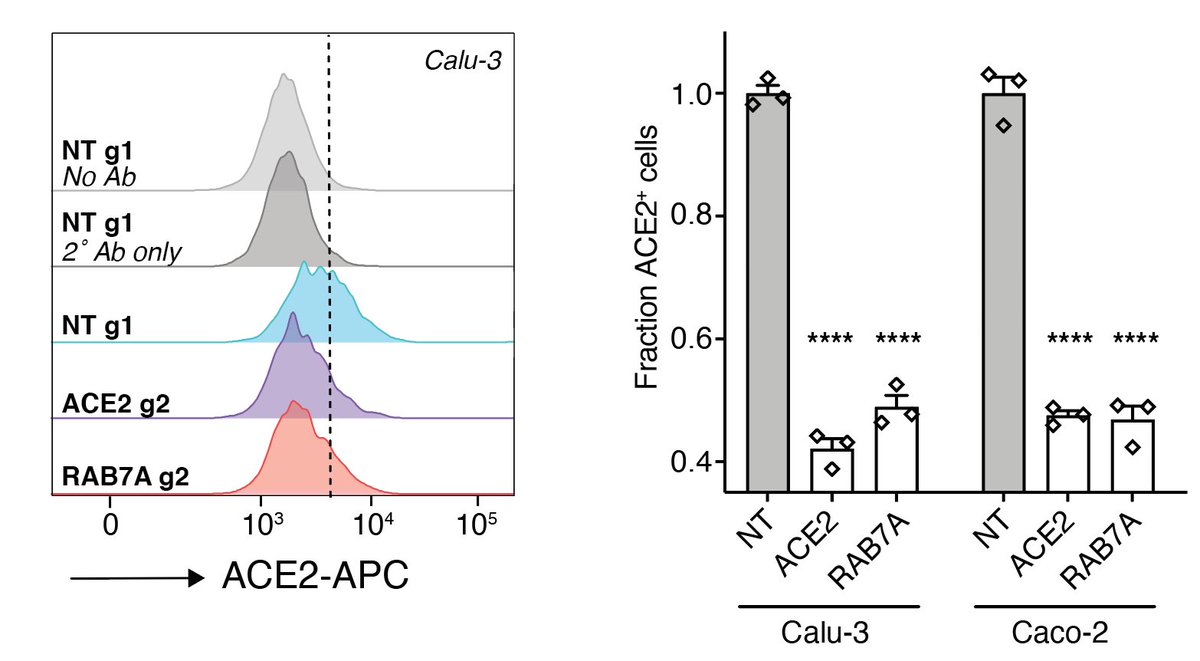
Since our cells were engineered to overexpress ACE2, we wondered whether we would see the same effect of RAB7A loss in other human cells. We do! It’s great to see that even genes identified using ACE2 overexpression cells translate to cells w/ endogenous ACE2. 
Moving forward, I am hopeful that some of the genes identified in our screen can help us find better therapies for COVID-19. Please get in touch if you are interested in helping push this along or support the next stages of our work.
On a related note, my friend @JohnDoench & @WilenLab recently published a beautiful screen using a monkey-targeting (!) CRISPR library with several different coronaviruses.
cell.com/cell/fulltext/…
cell.com/cell/fulltext/…
There are several excellent preprints of host factors from other groups. In particular, one study from #RongZhang and colleagues found many of the same protein complexes — lending further credence to these top-ranked hits:
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
Now, for the MOST important part of this thread, acknowledging the amazing scientists involved in this work. This was a true team effort and I am grateful to have been a part of this team.
First, this work was a wonderful collaboration between our group and the lab of Ben tenOever (@virusninja) at Mt. Sinai, who guided the work with a steady hand and creative ideas throughout.
We learned so much from Ben & his group. I am personally grateful for the opportunity he gave us to help contribute our skills to fighting the pandemic. (Also, he is indeed a virus ninja. 🧪🥷 Appropriate twitter handle! )
Another shout-out to Zharko & Tristan. Despite the distance between labs (a non-trivial issue during a pandemic), Zharko and Tristan were always in sync and doing their best to help each other out. Amazing NYC-wide teamwork in a challenging situation. Incredibly inspiring!
On our side, @ZDaniloski developed this idea from the earliest stages and his dedication & hard work was an inspiration to me. Zharko is just amazingly fun to do science with too. He also led the work on the Spike protein that I mentioned earlier in the thread.
I should mention that this project would not have been possible without several KEY folks. Special thanks to @we_harm, @emimitou, and @psmibert for cutting-edge single-cell work, @daisyhoagz for insightful amlodipine work, and @smaniatis for imaging wizardry.
During the review process, we unfortunately had to remove some super-exciting work integrating the genome-wide CRISPR screen with COVID-19 patient genetics (GWAS, eQTLs) that was led by @silvakasela and @tuuliel. Stay posted for updates from Tuuli & Silva soon!
BIG thank you also to @MatLegut, #LuLu, @egeller22, @odeddan, #BradRosenberg, and #HemaliPhatnani. Great people make great papers. These are great people. 🙌🙂
On the whole, the paper was also significantly improved by the reviewers — thank you to all of them. Also, I am grateful to funders of our lab who gave us the flexibility to use existing funds (@NIH and @DARPA) to support this new direction.
During this project, I was inspired seeing scientists of diverse backgrounds come together to attack a common problem. Early COVID efforts from my scientific mentors (bioengineer @zhangf & neuroscientist @SebastianSeung) convinced me to think about how I could contribute.
On a final note: We started this work back in March and it is a bit sad to see it published on a day with record daily infections in the US. 🙁 I am hopeful that innovation will help us move forward towards a brighter, COVID-free future. 👩🔬👨🔬✂️🧪💪
https://twitter.com/laurenpeikoff/status/1319588877556977665
Addendum: A few folks to acknowledge who helped us along the way: #TomManiatis for crucially connecting tenOever lab & our lab, @DrBenNeel for critical feedback on drug specificity, and #SylviaAdams & @Magdalenaplasil for sharing patient data. Thank you all! 👏
• • •
Missing some Tweet in this thread? You can try to
force a refresh