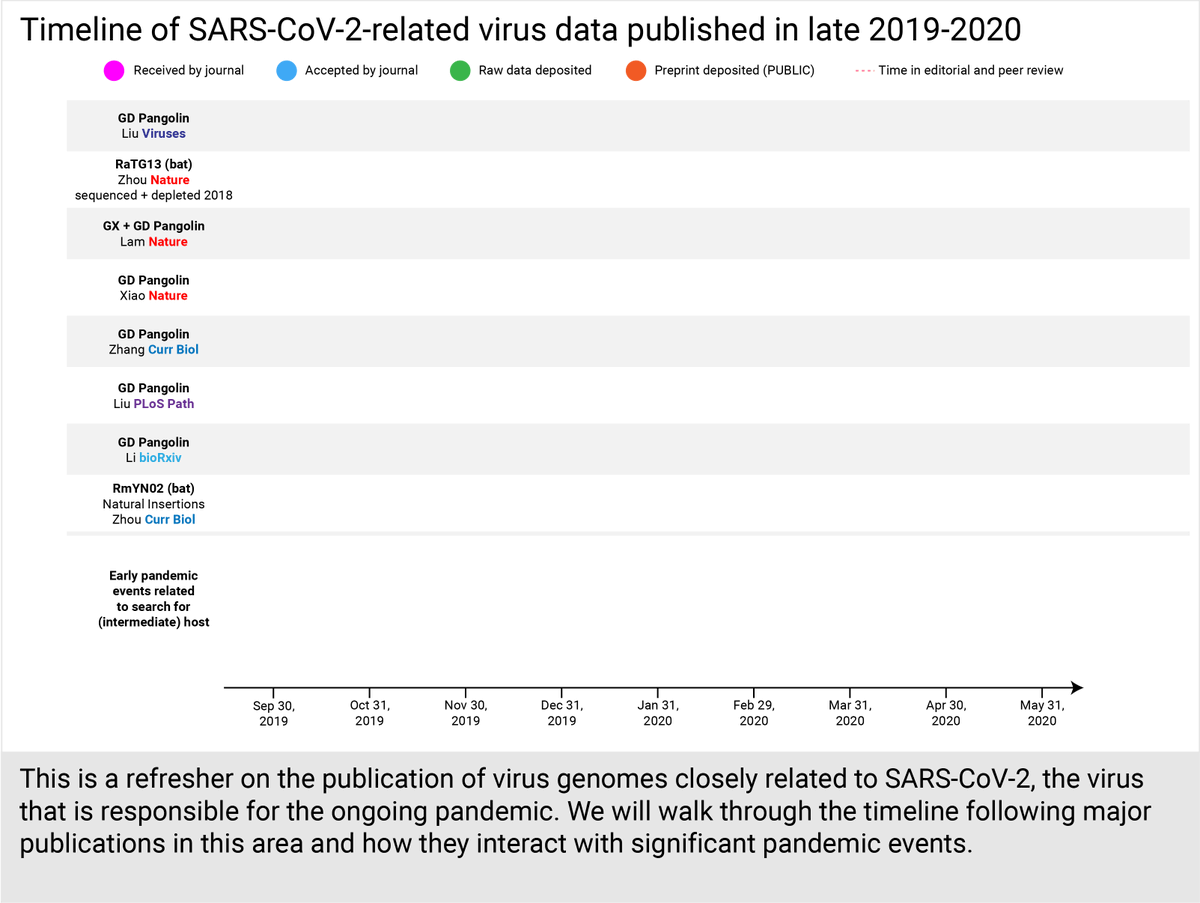
@AlexBerenson This is a question I've been asking as well. Where was RaTG13's data stored since 2018? Not in a public or password-restricted national database because even other Chinese groups only noticed the 99% match between SARS2 and the btCoV/4991 short sequence published in 2016 on NCBI.
@AlexBerenson Even scientists from the State Key Lab of Virology, Wuhan didn't have access to RaTG13 data (collected 2017-2018). In their Feb 5 paper, they lamented: "the BtCoV/4991 sequence was only partial and thus no comparisons can be made for the rest of genomes." tandfonline.com/doi/full/10.10…
@AlexBerenson Although the WIV publication that first named RaTG13 was put on bioRxiv on Jan 23, the raw data and genome sequence for RaTG13 were only deposited on a public database on Feb 13 and Mar 24, respectively. ncbi.nlm.nih.gov/nuccore/MN9965…
@AlexBerenson That means that up until that point, no one else appeared to have access to the RaTG13 data that had existed since at least 2017-2018 - as seen from the amplicon data (to fill gaps in the genome assembled from the raw data) quietly added to NCBI on May 19. ncbi.nlm.nih.gov/sra/SRX8357956
• • •
Missing some Tweet in this thread? You can try to
force a refresh