
Highlights from our #preprint on #transcription activation domains (ADs)!
-found all ADs in yeast TFs
-predicted yeast & human ADs
-tested binding of all ADs to #Mediator & #TFIID
-explained mechanism with structural modeling & kinetic experiments
(1/16)
biorxiv.org/content/10.110…
-found all ADs in yeast TFs
-predicted yeast & human ADs
-tested binding of all ADs to #Mediator & #TFIID
-explained mechanism with structural modeling & kinetic experiments
(1/16)
biorxiv.org/content/10.110…

We tested ~7500 53aa tiles covering all 164 yeast transcription factors (TFs) in an activation screen in vivo.
By controlling for fragment expression & sorting GFP activation into 8 bins, we get quantitative measurements with super high signal-to-noise. We found 150 ADs. (2/16)
By controlling for fragment expression & sorting GFP activation into 8 bins, we get quantitative measurements with super high signal-to-noise. We found 150 ADs. (2/16)

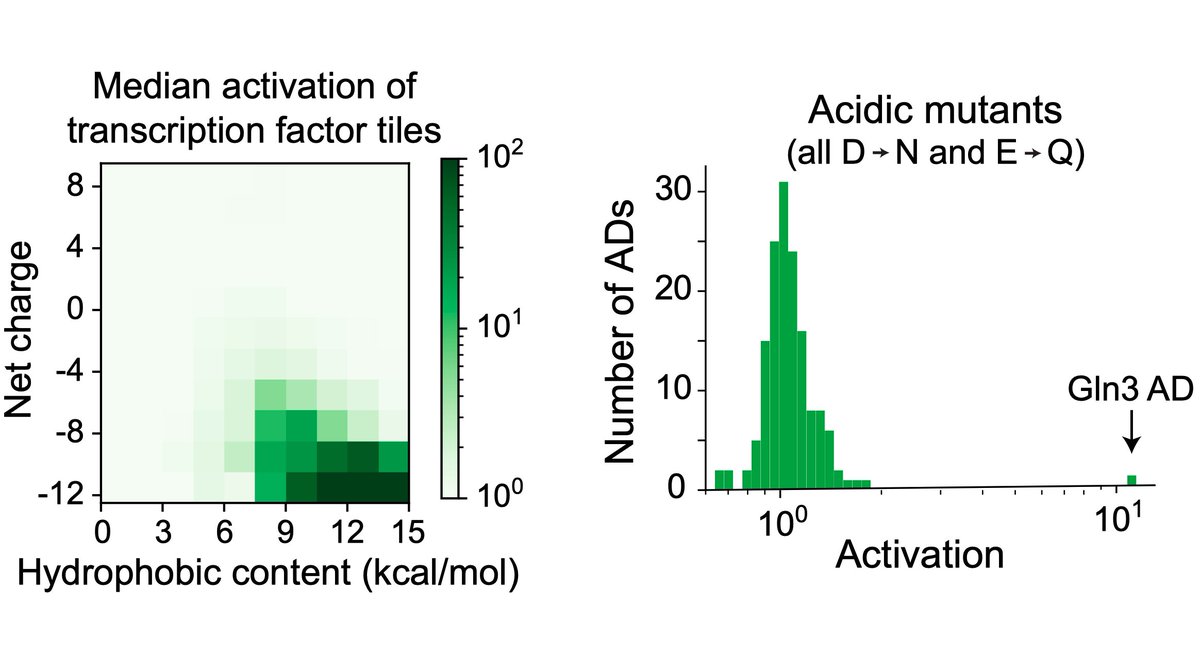
Tiles with lots of acidic & hydrophobic residues activated most.
But there are conflicting reports on whether acidic residues are important, so we mutated all the acidic residues in each AD. Besides one interesting case, no mutants activated.
So all yeast ADs are acidic! (3/16)
But there are conflicting reports on whether acidic residues are important, so we mutated all the acidic residues in each AD. Besides one interesting case, no mutants activated.
So all yeast ADs are acidic! (3/16)
We also took 8 known ADs and titrated the amount of negative charge and hydrophobicity by removing more and more acidic or aromatic residues.
Activation was gradually and then completely eliminated, and was most related to the number rather than the position of mutations. (4/16)
Activation was gradually and then completely eliminated, and was most related to the number rather than the position of mutations. (4/16)

We trained the Predictor of Activation Domains using Deep Learning in Eukaryotes (PADDLE), which predicts the location and strength of ADs in new proteins.
PADDLE also predicts the activation of mutants and homologs very accurately. (5/16)
PADDLE also predicts the activation of mutants and homologs very accurately. (5/16)

Classic experiments showed that ADs function even when transferred between yeast and human.
We used PADDLE to identify hundreds of acidic ADs in human TFs and virus proteins.
We tested 25 of them using a luciferase assay in human cells and 92% of them activated! (6/16)
We used PADDLE to identify hundreds of acidic ADs in human TFs and virus proteins.
We tested 25 of them using a luciferase assay in human cells and 92% of them activated! (6/16)

PADDLE also identifies the short, activating core regions of each AD. They are extra acidic & hydrophobic.
We tested >2000 mutants of 28 core ADs and figured out that only a few require helical folding and most activate just as strongly when their sequence is scrambled! (7/16)

We tested >2000 mutants of 28 core ADs and figured out that only a few require helical folding and most activate just as strongly when their sequence is scrambled! (7/16)


To understand how such diverse sequences can activate, we tested their binding to coactivator proteins.
We expressed the library of TF tiles as peptides tagged by their mRNA sequence, bound it to bait protein, and sequenced the input and bound fractions. (8/16)
We expressed the library of TF tiles as peptides tagged by their mRNA sequence, bound it to bait protein, and sequenced the input and bound fractions. (8/16)

We tested binding of TF tiles to the Med15 subunit of #Mediator and to #TFIID and saw clear binding domains.
73% of ADs bound to Med15.
Only 18% bound TFIID, and all of these bound Med15. (9/16)
73% of ADs bound to Med15.
Only 18% bound TFIID, and all of these bound Med15. (9/16)

Activation and Med15-binding strength were correlated across wt TF tiles.
We took the aromatic mutants of 7 ADs and tested their Med15 binding too. Activation and binding were tightly correlated.
So the same hydrophobic features underlie activation and Med15 binding. (10/16)

We took the aromatic mutants of 7 ADs and tested their Med15 binding too. Activation and binding were tightly correlated.
So the same hydrophobic features underlie activation and Med15 binding. (10/16)


How can Med15 bind diverse ADs?
Med15 has 4 activator-binding domains (ABDs). We modeled binding of 28 core ADs to two ABDs.
Shape complementarity was weak. ABDs were bad at burying hydrophobic AD residues.
Consequence: all ADs bound in many poses, aka fuzzy complex. (11/16)
Med15 has 4 activator-binding domains (ABDs). We modeled binding of 28 core ADs to two ABDs.
Shape complementarity was weak. ABDs were bad at burying hydrophobic AD residues.
Consequence: all ADs bound in many poses, aka fuzzy complex. (11/16)

2nd consequence: ABDs aren't picky about what hydrophobic residue binds.
e.g. we plotted positions of AD residues on ABD surface. Leu and Phe had no specific binding pockets, they bound the same areas.
This is why AD sequences can be so diverse! (12/16)
e.g. we plotted positions of AD residues on ABD surface. Leu and Phe had no specific binding pockets, they bound the same areas.
This is why AD sequences can be so diverse! (12/16)

This fuzzy binding is weak, so high affinity comes from high valence
We measured kinetics of Mediator complex recruitment to the TF Gcn4 on DNA. Affinity is high (14nM) b/c Gcn4 has 3 core ADs & forms a dimer
More Gcn4 DNA motifs -> longer binding, even higher affinity. (13/16)
We measured kinetics of Mediator complex recruitment to the TF Gcn4 on DNA. Affinity is high (14nM) b/c Gcn4 has 3 core ADs & forms a dimer
More Gcn4 DNA motifs -> longer binding, even higher affinity. (13/16)

But even 250x excess of competitor Gcn4 didn't slow Mediator binding to Gcn4 AD on DNA.
So high-valence binding allows rapid, dynamic exchange.
Dynamism is a key feature! eg Mediator must be released after transcrip initiation. Explains why this binding mode is used. (14/16)
So high-valence binding allows rapid, dynamic exchange.
Dynamism is a key feature! eg Mediator must be released after transcrip initiation. Explains why this binding mode is used. (14/16)

We predicted/confirmed ADs in all major yeast coactivator complexes. Many are disordered, so should be accessible
eg several in cohesin. Responsible for known interaction with Mediator?
These ADs probably drive phase separation. But we think dynamism is the key feature. (15/16)
eg several in cohesin. Responsible for known interaction with Mediator?
These ADs probably drive phase separation. But we think dynamism is the key feature. (15/16)

Big thanks to Roger Kornberg for letting me pursue ideas & use/develop many techniques w/ full independence!
Great working w/ @bentyeh (library design & PADDLE), @raphaeljlt & Ron Dror (modeling), Cynthia Hao & Jordan Feigerle (experiments), and @erezaterez (sequencing)! (16/16)
Great working w/ @bentyeh (library design & PADDLE), @raphaeljlt & Ron Dror (modeling), Cynthia Hao & Jordan Feigerle (experiments), and @erezaterez (sequencing)! (16/16)
ADs are domains on transcription factor proteins that activate transcrip when at a promoter/enhancer
Function is conserved yeast to human but poorly conserved in sequence, so annotations are sparse!
Has been hard to define a mechanism unifying how diverse AD sequences activate
Function is conserved yeast to human but poorly conserved in sequence, so annotations are sparse!
Has been hard to define a mechanism unifying how diverse AD sequences activate
• • •
Missing some Tweet in this thread? You can try to
force a refresh


