
Excited to share our paper on a fast (<40 min. TAT), cheap (<$6/sample), accessible (no special equipment, sous vides welcome), sensitive (LoD ~25 copies/uL) SARS-CoV-2 LAMP-based #covidtest, published in #OFID doi.org/10.1093/ofid/o…. My first 🧵(1/n)
Preface: V. grateful for the collaboration between @mghpathology (Eric Rosenberg, Graham McGrath, Joe Lennerz, John Branda, and 🏆 clin micro lab staff), @MassGeneralEM (Benjamin White and 👏ED staff), Harvard Med (@brianrabe6, Connie Cepko) & @NEBiolabs (@nathan_a_tanner) (2/n)
We started this work in early March. The COVID surge was coming but clinical micro labs faced countless shortages (real-time PCR machines, RNA extraction kits, master mix, media). Borrowing from generous research labs only got us so far. Was there an alternative to qPCR? (3/n)
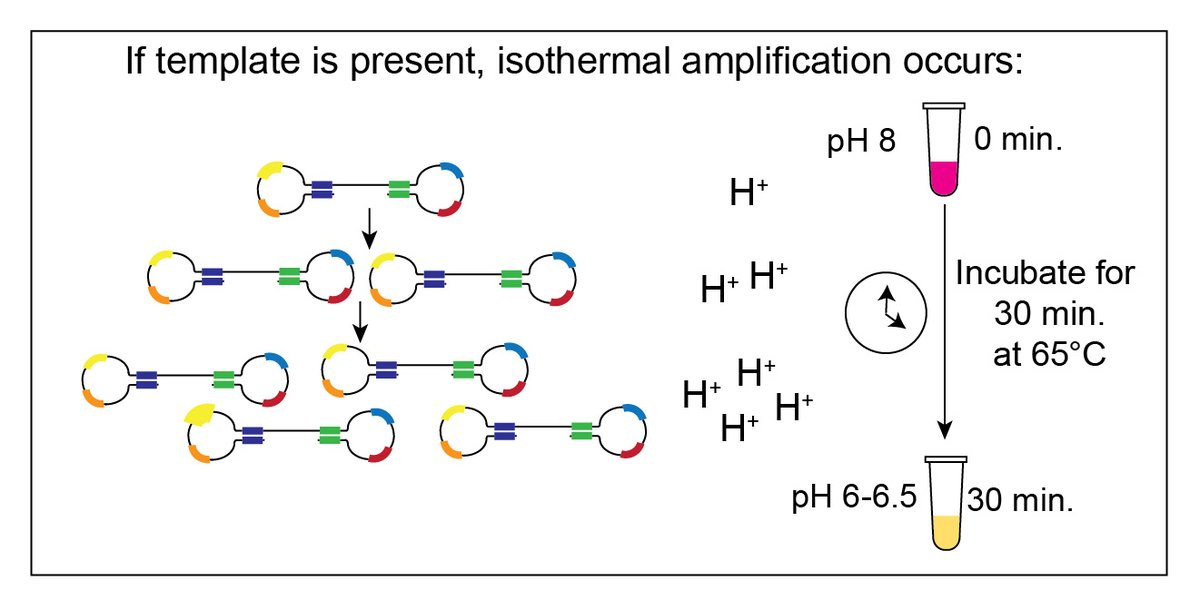
Colorimetric RT-LAMP is as simple as 🧬 tests get.
1. Mix sample, master mix, LAMP primers
2. Incubate 30 min. @ 65C for reverse transcription & amplification
3. Visual read out (pink=neg, yellow=positive; pH based).
No thermocyclers, fluorescent readers, lat flow strips. (4/n)
1. Mix sample, master mix, LAMP primers
2. Incubate 30 min. @ 65C for reverse transcription & amplification
3. Visual read out (pink=neg, yellow=positive; pH based).
No thermocyclers, fluorescent readers, lat flow strips. (4/n)
@nathan_a_tanner's preprint (doi.org/10.1101/2020.0…) showed promising SARSCoV2 detection from extracted RNA w/ colorimetric RT-LAMP. But RNA extraction kits were 💰 & in limited supply. Even w/o COVID, spin columns are painful & slow at large-scale. Could we skip RNA extraction?
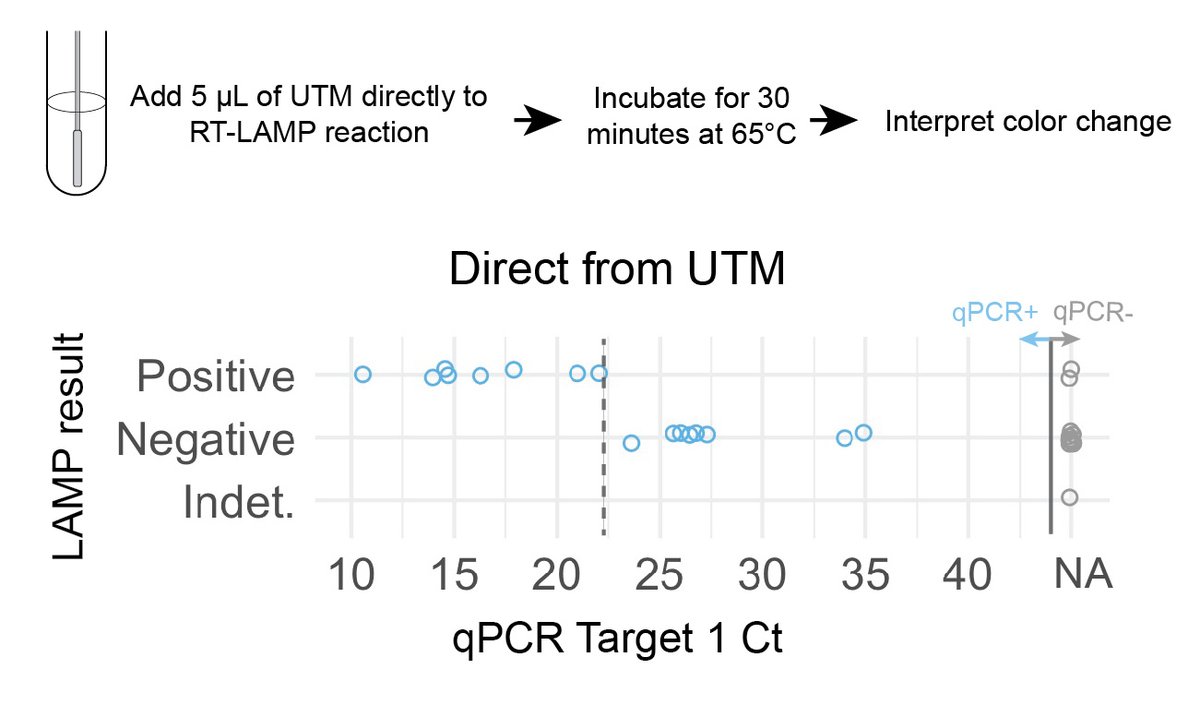
We tried direct-from-UTM detection first. NP swabs collected in UTM ➡️ directly added UTM to LAMP rxn. How much UTM could we add per rxn? W/ ⬆️ sample input volumes, the buffer and phenol red in UTM interfered with color change. Could only add ~1 µL of UTM to each LAMP rxn. (6/n)
We then tested 16 qPCR+, 15 qPCR- NP specimens with direct-from-UTM LAMP. Unfortunately, could only detect cycle thresholds (Ct) <23 (~3,000 copies/µL for us). Detects the most highly infectious patients, but misses many +s. Two false +s, color change was hard to interpret.(7/n) 
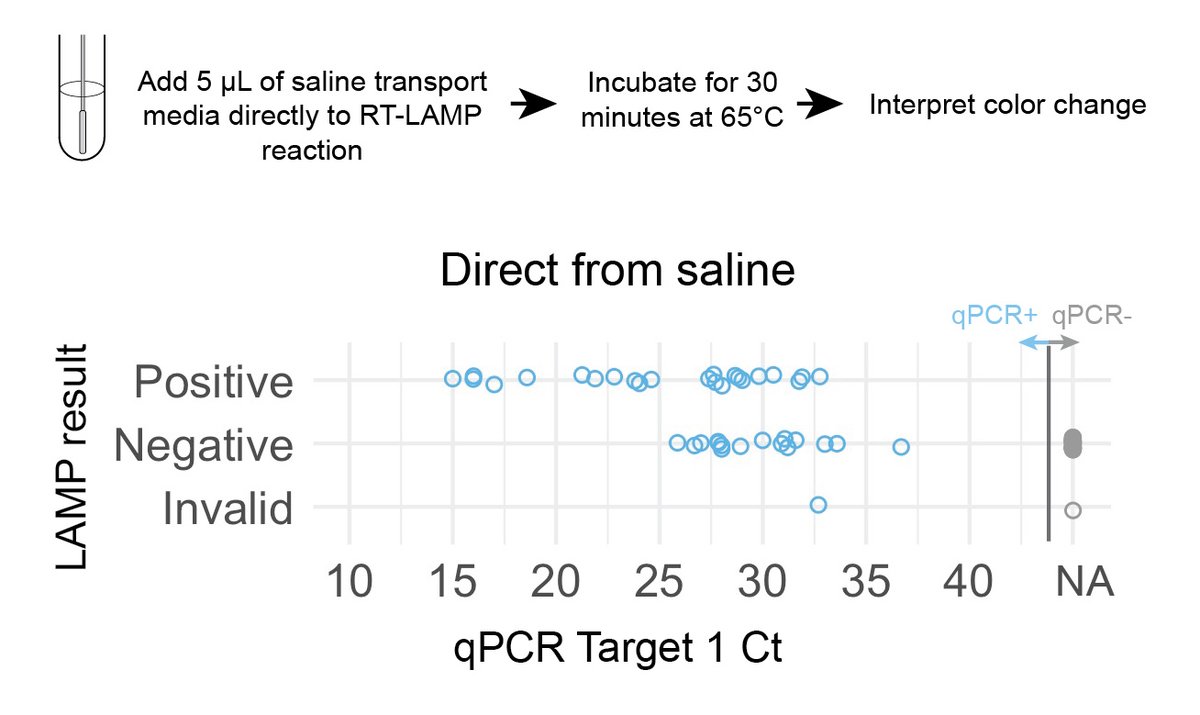
What if we collected NP swabs in sterile saline instead of UTM?
✔️ No buffer or phenol red to interfere w/ colorimetric read-out.
✔️ No supply shortage of saline.
We found we could add larger sample volumes (5-10 µL) to LAMP rxn w/o interference. (8/n)
✔️ No buffer or phenol red to interfere w/ colorimetric read-out.
✔️ No supply shortage of saline.
We found we could add larger sample volumes (5-10 µL) to LAMP rxn w/o interference. (8/n)
Direct-from-saline testing could detect samples with Cts as high as 33, but still missed many qPCR+ samples (assay sens ~59%). Again, great for detecting highly infectious people but still missed true positives. Crisper color changes with saline than UTM. No false +s. (9/n) 
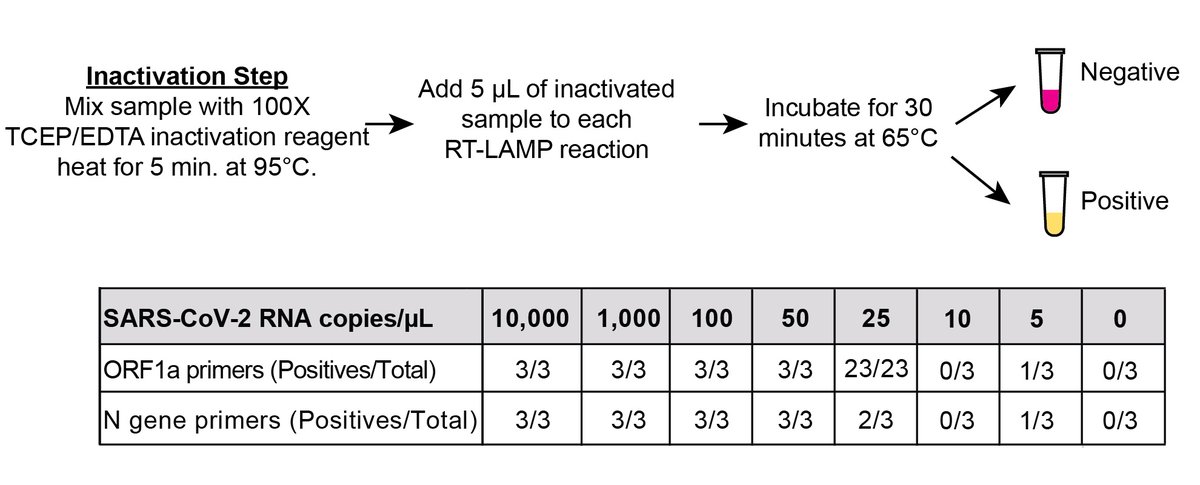
Nearly stopped here, until we met @brianrabe6 & Dr.Cepko, who optimized a chemical (TCEP/EDTA)+🔥 (95C for 5 m.) protocol to inactivate endogenous RNases in specimens & release/lyse virions (doi.org/10.1073/pnas.2…), based on @cambearon @CatherineFreije @sabetilab's HUDSON (10/n)
We could finally perform true LoD studies by spiking naked control RNA into TCEP/EDTA/heat inactivated NP specimens (previously, endogenous RNases chewed up spiked-in RNA immediately). LoD was ~25 copies/µL of sample, huge improvement! Would it work w/ clinical samples?(11/n) 
We incorporated that 5 min. chemical/heat inactivation protocol into a rigorous testing procedure that tested each sample w/ 2 separate LAMP primer sets (ORF1a & N gene) + an internal control (human actin), in parallel w/ + and – external controls. (12/n) 
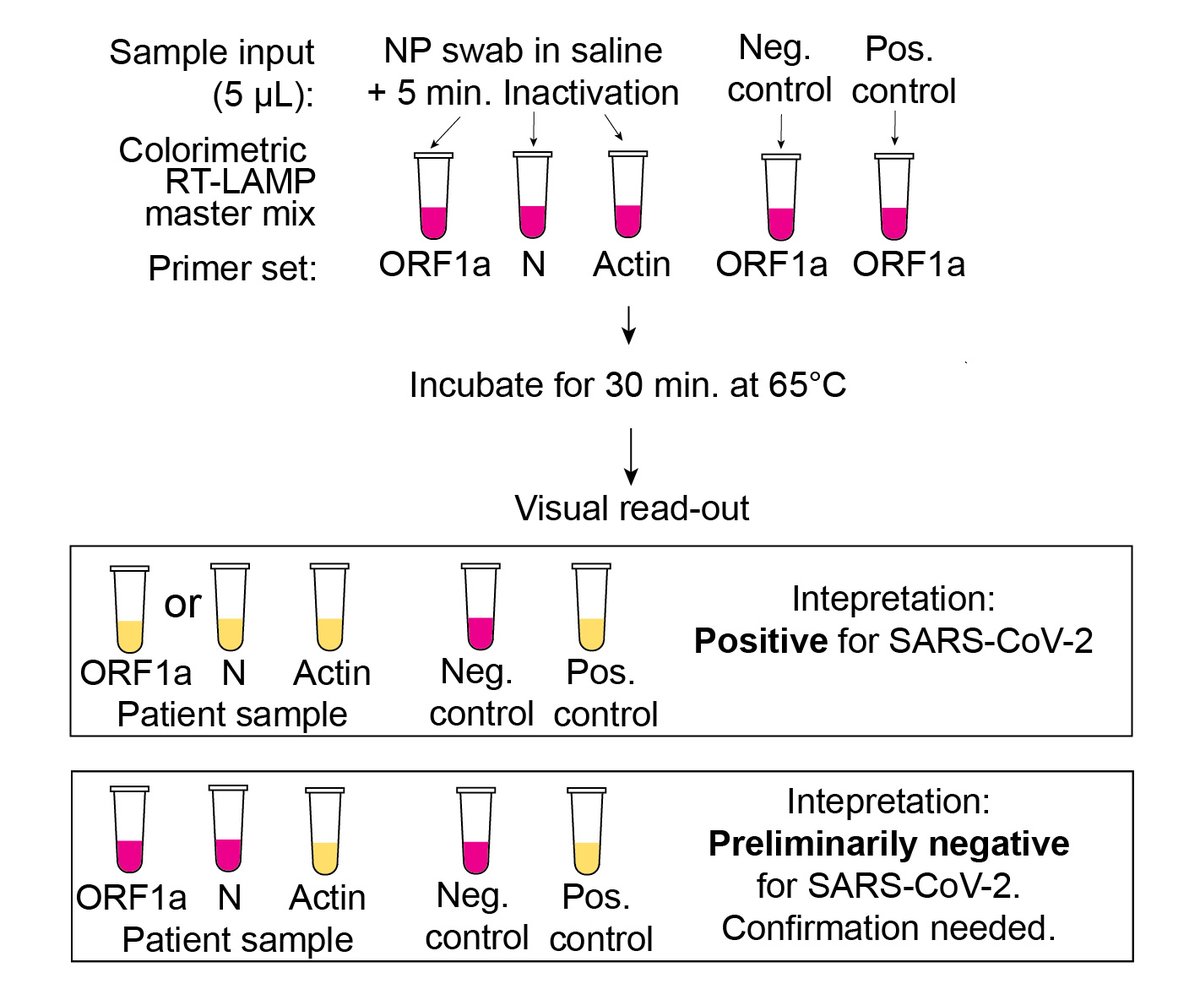
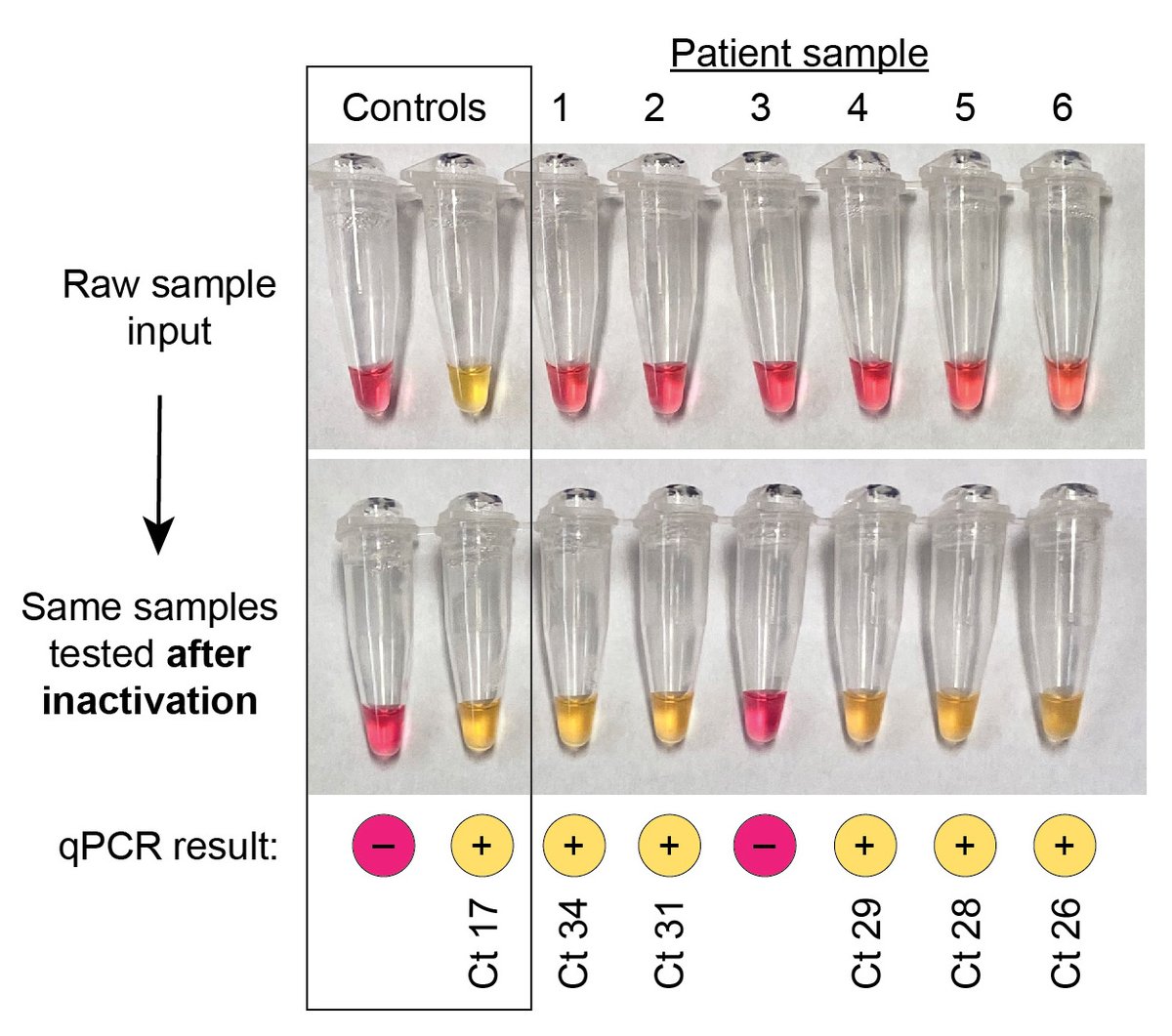
Inactivation pre-LAMP had huge benefit: clinical samples with Ct 26-34 that were previously LAMP- but PCR+ were now LAMP+ too (top= 5 µL saline added directly to LAMP rxn, bottom = 5 µL of same sample tested after 5-min inactivation step).(13/n) 
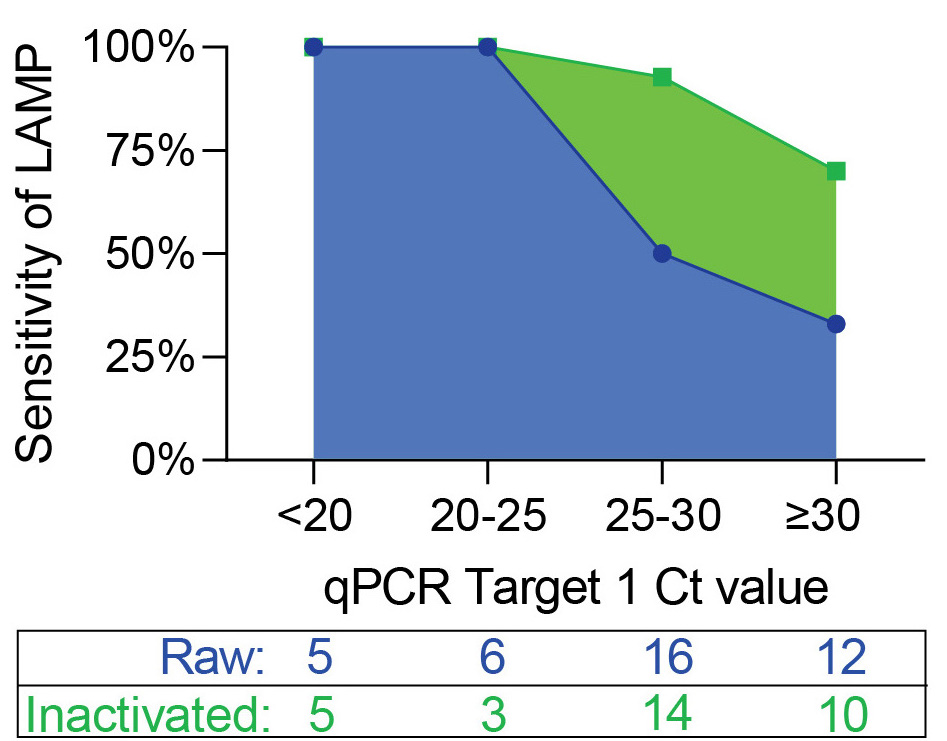
The simple inactivation step boosted sens. when we tested 32 qPCR+ and 30 qPCR- samples (Blue = raw samples (just added saline directly to LAMP rxn) vs. Green = inactivated samples added to LAMP rxn.) Found that inact.+LAMP had 87% sens & 100% spec in real-world sample set.(14/n) 
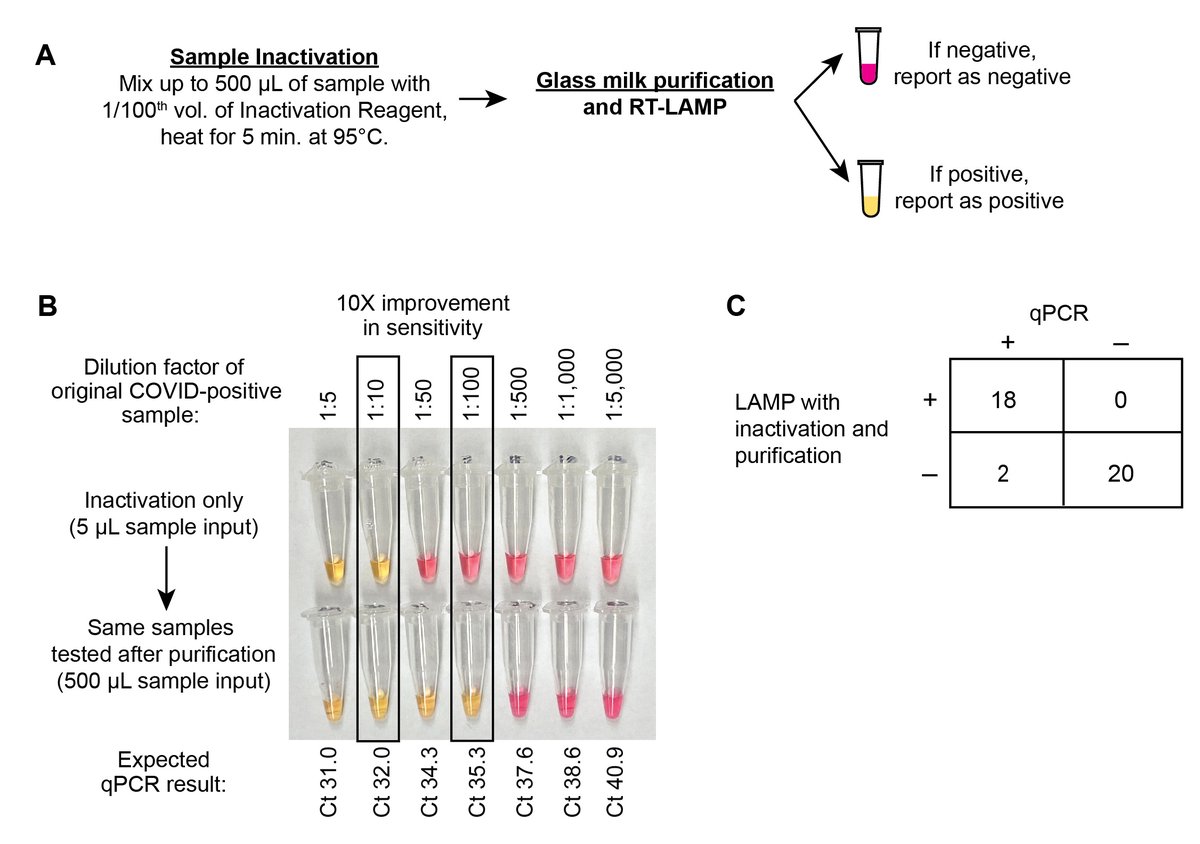
Finally, we tested a cheap ($0.07/sample) “glass milk” (i.e. silica particle) concentration step by @brianrabe6 & Dr.Cepko. Concentrates 500 µL of inactivated sample into 1 LAMP rxn w/10x improvement in LoD, ~= to qPCR. Picked up some v. low VL samples. 100% specific (15/n) 
In summary, we hope these simple protocols will improve access to rapid #covid19 testing, especially in resource-limited settings. Widely accessible, can order everything off-the-shelf. Only need water bath, pipette, freezer, PPE. (16/n)
Many others, incl. a global LAMP consortium started by @mason_lab, have similarly shown the ability to quickly and accurately detect SARS-CoV-2 using LAMP in a variety of ways. @ColorGenomics 1st w/ FDA EUA from RNA. @NEBiolabs now sells a colorimetric RT-LAMP kit,too.(17/n)
Limitations: no quantitation (no Ct value), relies on visual interpretation, needs -20C storage of master mix (though could lyophilize). Careful timing of incubation is important (non-specific amplification can happen after 30 min). (18/n)
Questions/comments welcome. Thanks for reading! (fin)
• • •
Missing some Tweet in this thread? You can try to
force a refresh


