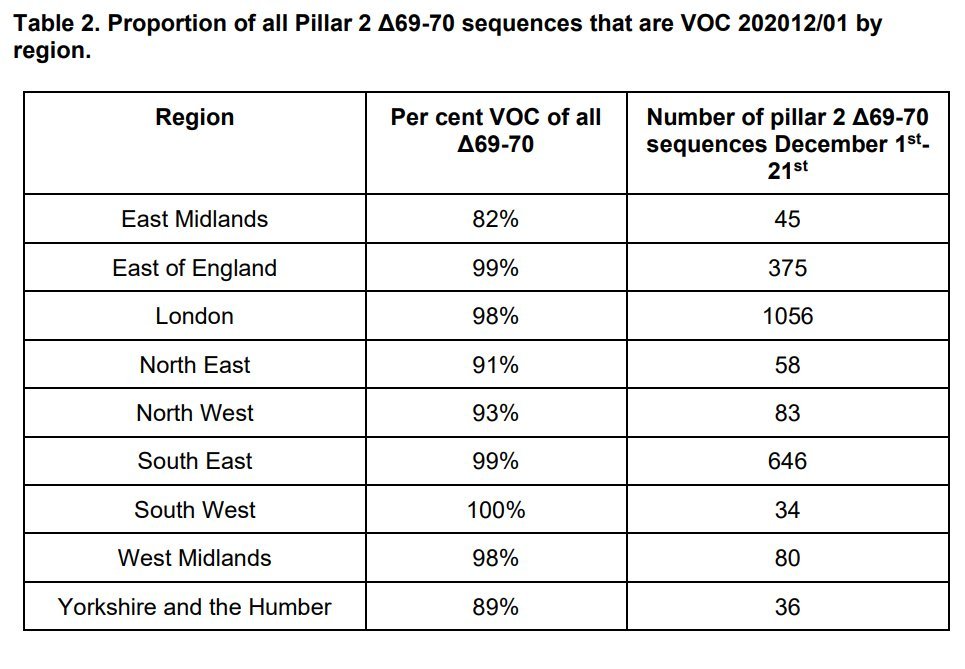
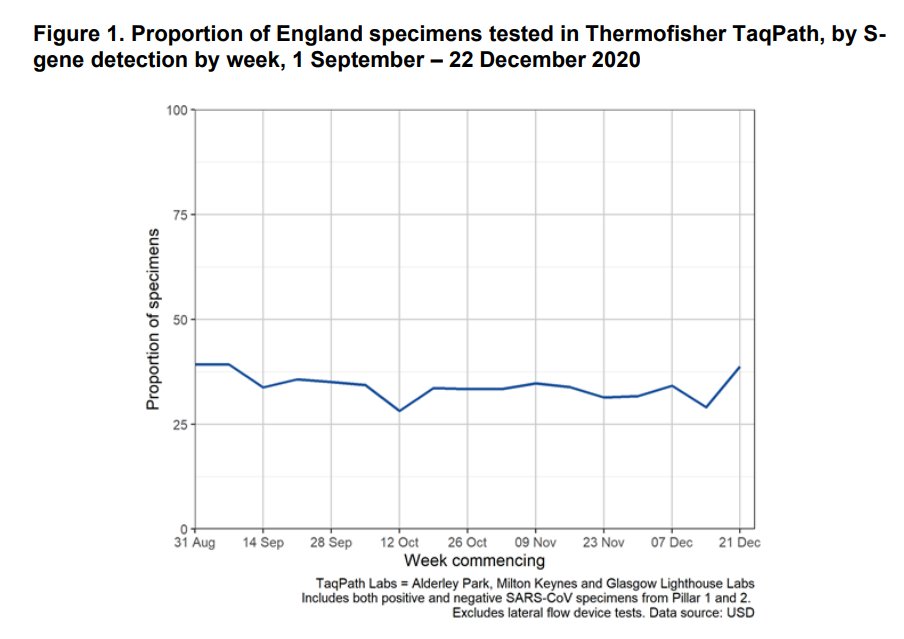
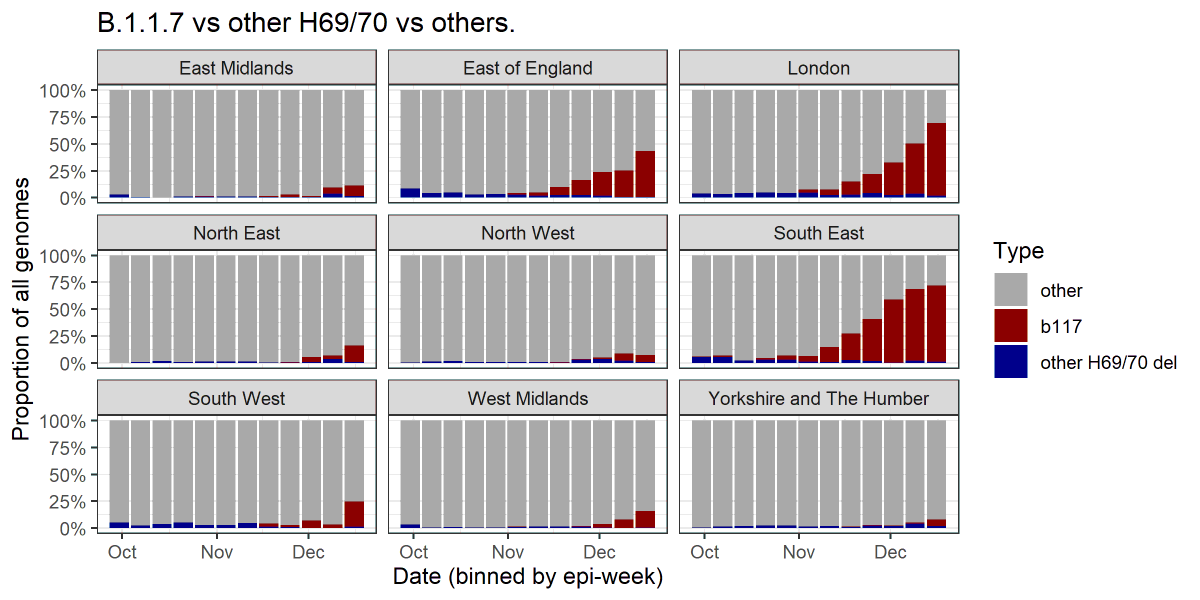
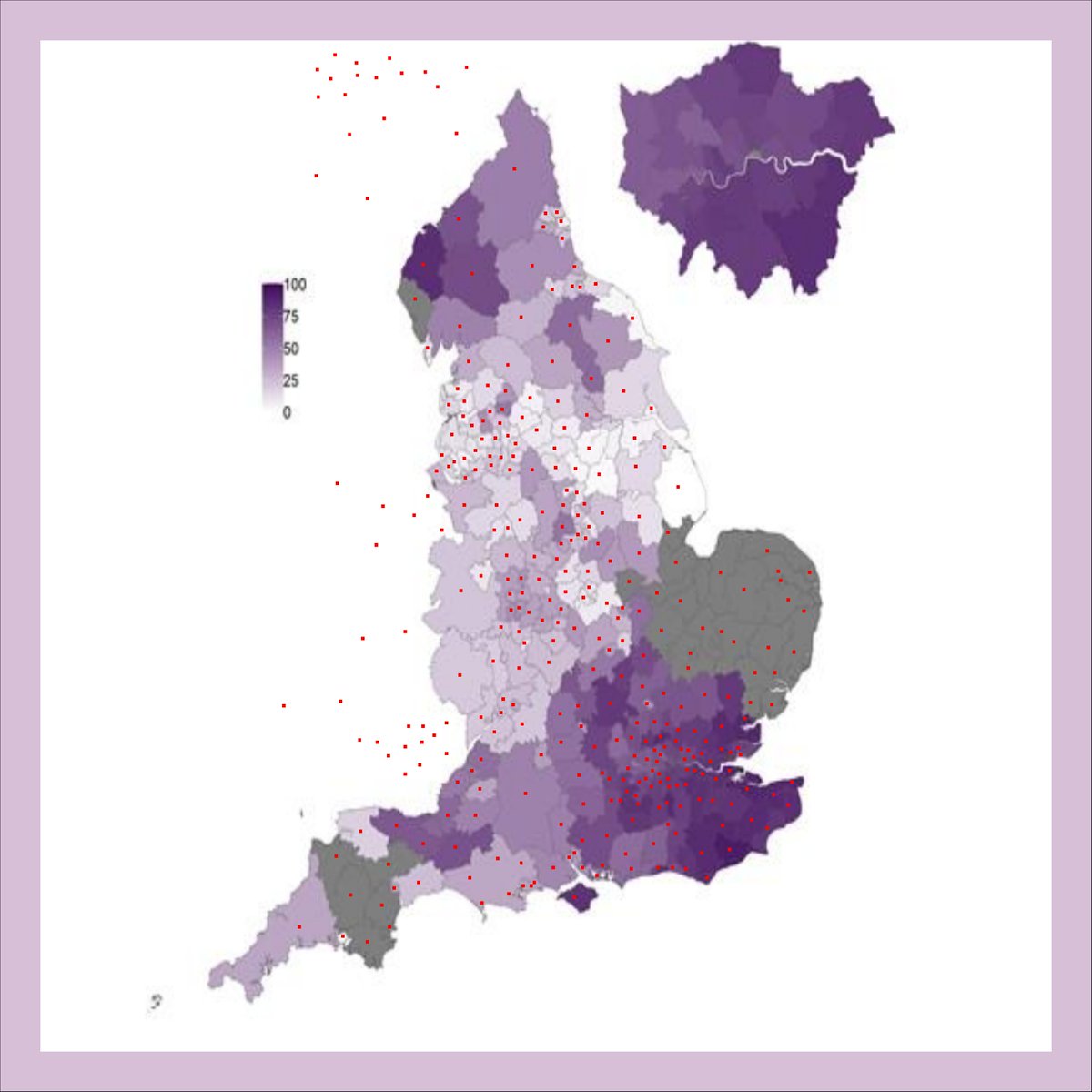
Had fun™ trying to extract S-dropout rates from the graph in the PHE report using an automated eyedropper. Take the values with a big pinch of salt!
theosanderson.github.io/adhoc_covid/ph…
theosanderson.github.io/adhoc_covid/ph…
CSV output with the source: github.com/theosanderson/…
And we can then compare this to the rate of growth in case figures week-on-week. (N.B. the already published analysis from PHE and others does this in much better more sophisticated ways, it's just interesting to explore) 
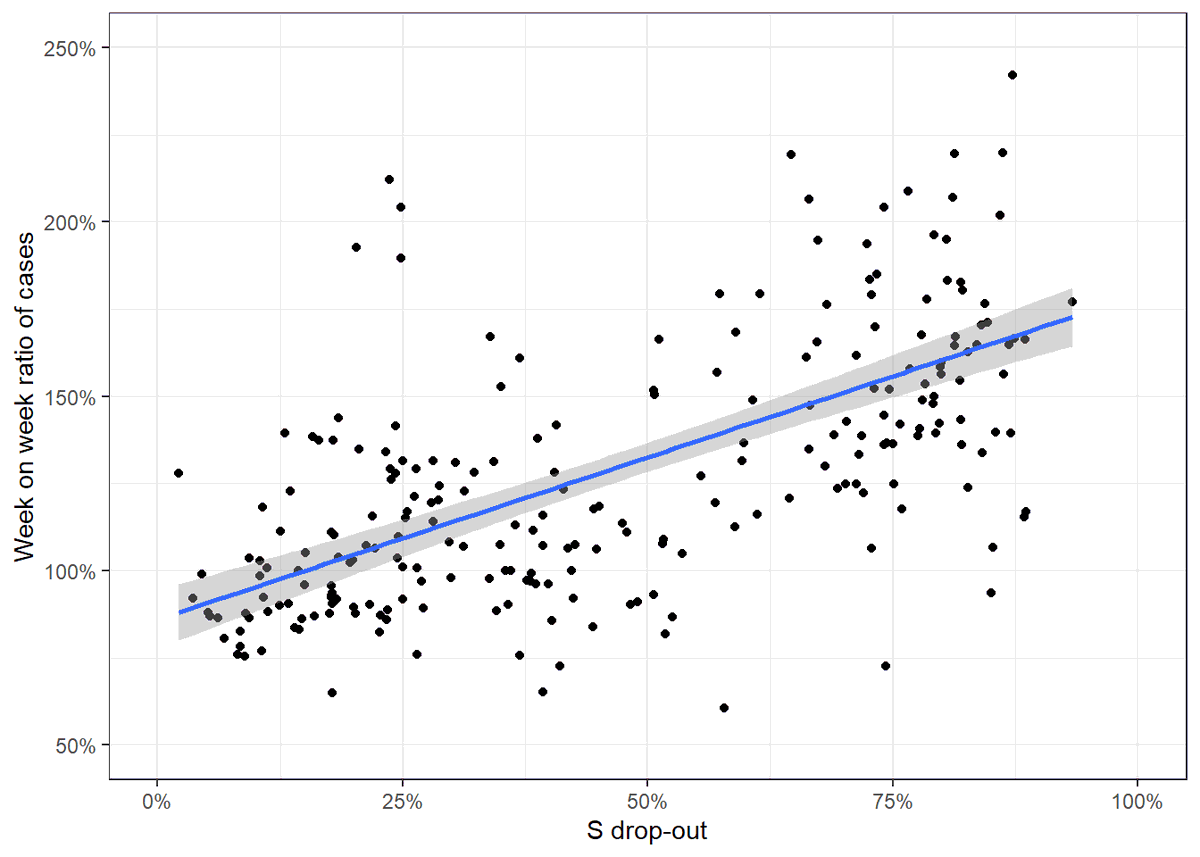
Actually looks like this went pretty well
• • •
Missing some Tweet in this thread? You can try to
force a refresh