
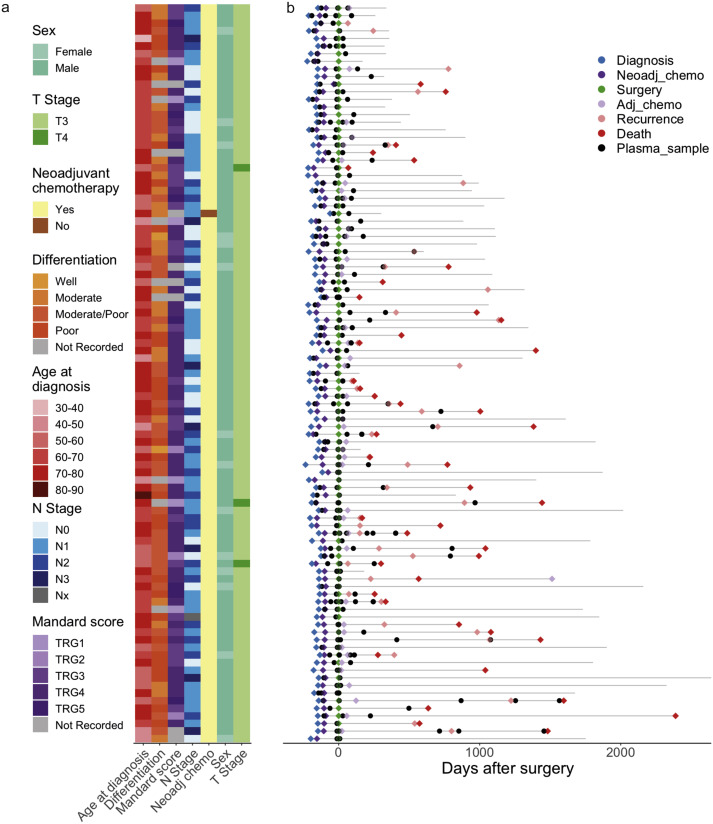
Here is a tweetorial from our latest publication in @Annals_Oncology about longitudinal tracking of esophageal adenocarcinoma. #OesophagealCancer #EsophagealCancer annalsofoncology.org/article/S0923-… 1/8
We sequenced 245 plasma samples from 97 patients with oesophageal adenocarcinoma using a 77 gene pan-cancer ctDNA panel. 2/8 
Variants derived from previously characterised driver oesophageal adenocarcinoma genes had a significantly higher VAF than variants from other genes, indicating selection. 3/8 
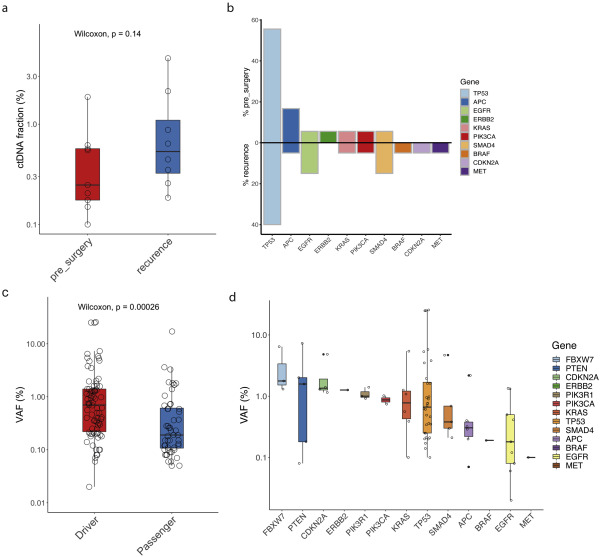
Peripheral blood cell samples were also sequenced for 78/97 patients. CHIP mutations were identified in 23% of cases, longitudinal tracking of CHIP variants suggested these variants were dynamic over time. 4/8 

We found patients that were ctDNA positive post-surgery had a significantly poorer survival than ctDNA negative patients, and the elimination of CHIP variants improved the positive predictive value. 5/8 
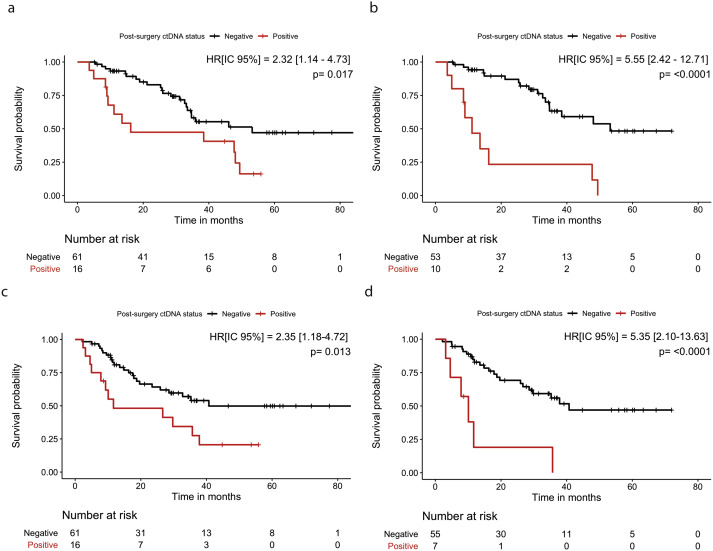
In summary, we demonstrate in a large, national, prospectively-collected dataset that ctDNA in plasma following surgery for EAC is prognostic for relapse. Inclusion of peripheral blood cell samples can reduce or eliminate false positives from CHIP. 6/8
In the future, post-operative ctDNA could be used to risk stratify patients into high- and low-risk groups for intensification or de-escalation of adjuvant chemotherapy. 7/8
Many thanks to our funders and patients who participated within the #OCCAMS consortium. The study was carried out by our brilliant PhD student Emma Ococks, medical oncologist @LizzySmyth1, postdocs @AFrankell and @neus_snows and others @MRC_CU, @Cambridge_Uni, @CRUKresearch. 8/8
@threadreaderapp unroll please
• • •
Missing some Tweet in this thread? You can try to
force a refresh



