Did you know #lipids control cell identity? Yes, they do! Happy to share our first preprint on #singlecell #lipidomics in collaboration with @gio_dangelo and a fabulous team led by @CapolupoLaura and Irina Khven. biorxiv.org/content/10.110… 
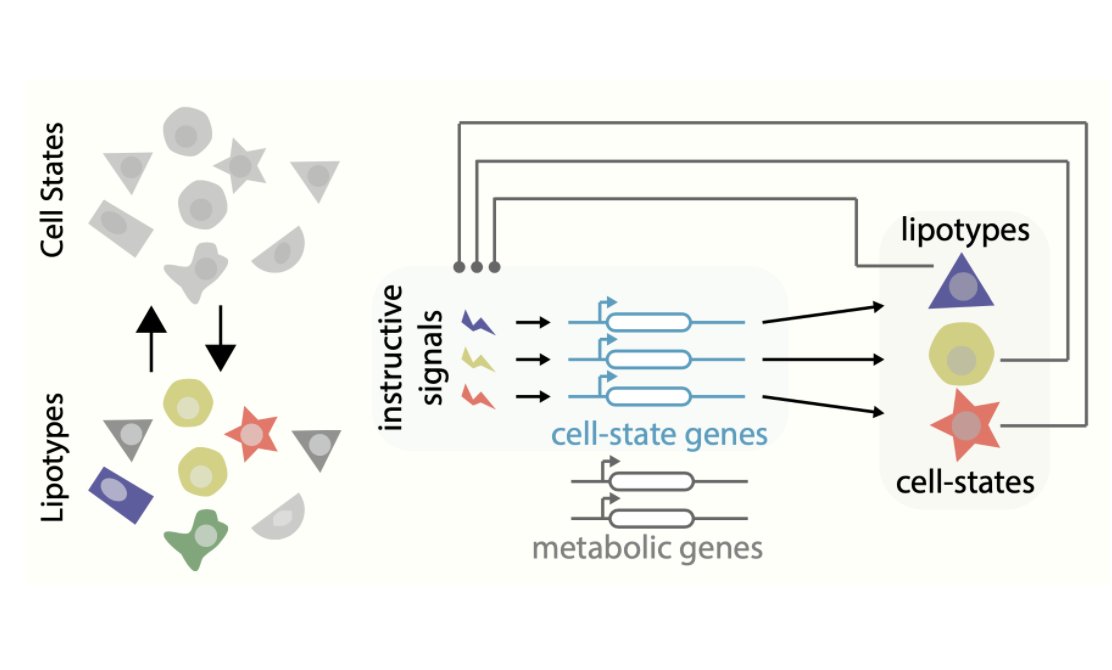
We used #MALDI imaging mass spectrometry to measure the single-cell lipidomes of hundreds of individual human dermal #fibroblasts. We identify specific lipid metabolic pathways that display cell-to-cell variability. Unexpectedly, single cells clustered by lipid composition! 

It took a lot of experiments to convince us, but now we can say it confidently: there is such a thing as a #lipotype!! First of all, toxin-based lipid staining validated the existence of the different populations of dermal fibroblasts both in vitro and in vivo. 
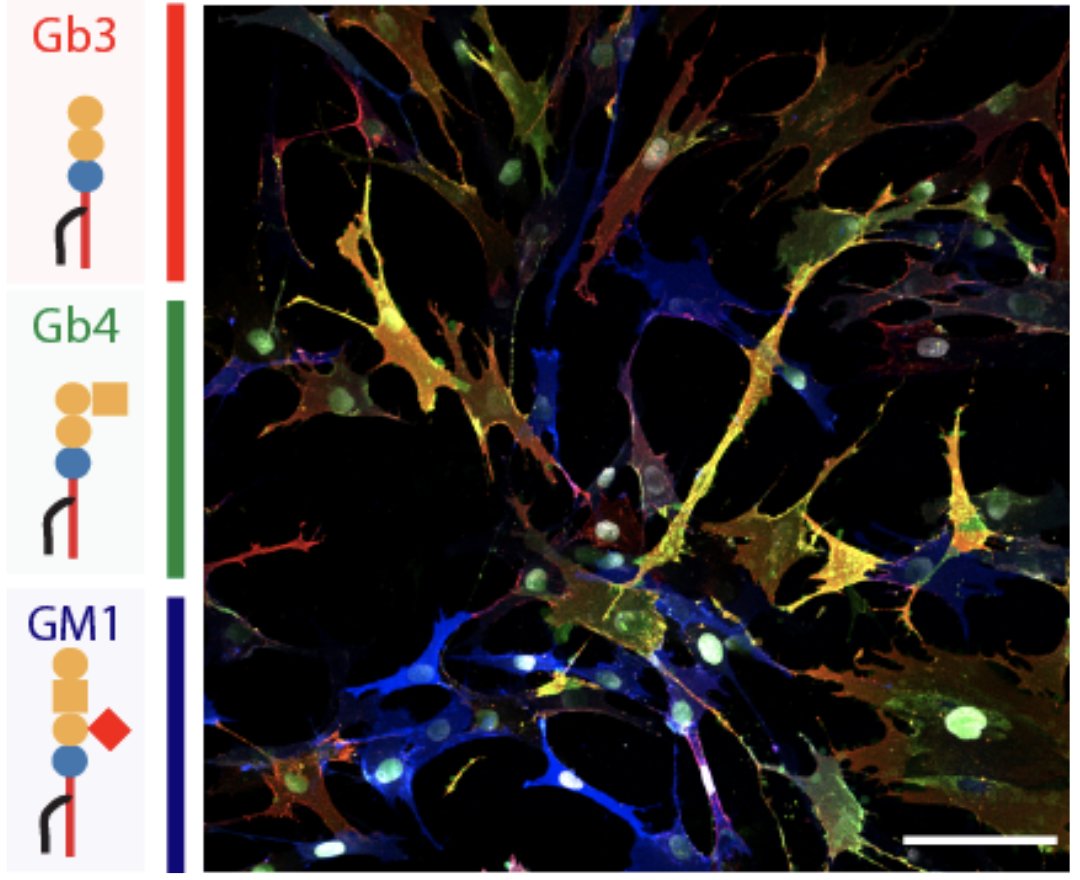
Second, by coupling #scRNAseq with #scLipid analysis (yes, a new flavor of #multiomics !), we found that there is a correspondence between discrete lipid metabolic conformations and characteristic fibroblast transcriptional states. 
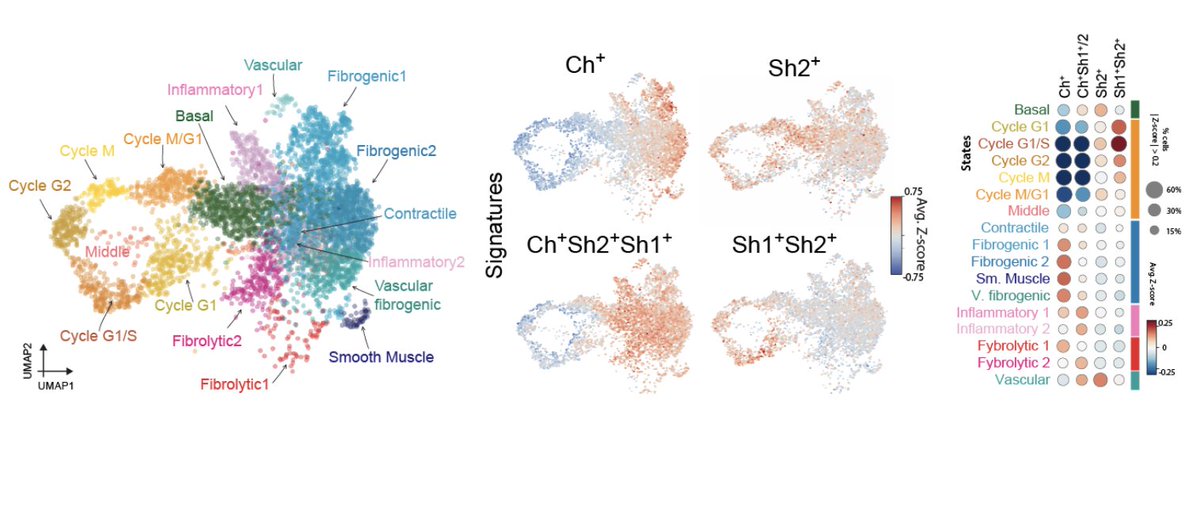
However, #lipotypes are not just the “metabolic shadow” of transcriptional heterogeneity. Enzymes that synthesize lipids are expressed homogeneously across cells and do not correlate with cell-state markers. The correspondence must come from post-transcriptional regulation. 

If one wipes out the class of lipids that make up lipotypes, fibroblasts change cell state! This happens through an imbalance of the FGF-TGFb pathways. Manipulating specifically single metabolic reactions, we saw again the same result: fibroblasts change their state. 

More experiments and analyses confirmed that FGF/TGF signaling contributes to determining cell lipotypes. Interesting to see how signaling and metabolism can be wired-up in self-contained circuits that control cell identity. 

We have been thrilled to observe a biologically-relevant function for cell-to-cell metabolic heterogeneity and are excited to see how #multiomics single-cell analysis can drive us towards uncharted domains of biology.
If proof was needed to the few still #singlecellSkeptics out there, here you have it, and I can say it out loud: #singlecell analyses can be used to learn mechanistic aspects of biology!
Of course, @GiO__DAngelo is the right Twitter handle 😅
• • •
Missing some Tweet in this thread? You can try to
force a refresh





