
I'm very excited to share my very first paper!!💜
In our work in @ActaBio we propose a more sophisticated way of validating #3D #TractionForceMicroscopy methods and we apply our novel inverse method to an in vitro model of #angiogenesis (1/10)
👇👇🧵
sciencedirect.com/science/articl…
In our work in @ActaBio we propose a more sophisticated way of validating #3D #TractionForceMicroscopy methods and we apply our novel inverse method to an in vitro model of #angiogenesis (1/10)
👇👇🧵
sciencedirect.com/science/articl…
Typically, TFM methods are validated under simplified scenarios (using simplified cell geometries, arbitrarily choosing force exertion points, or bypassing image processing steps). Here, we designed a simulation platform that is as close as possible to a real case. (2/10)
We used the geometry of a real angiogenic sprout via confocal microscopy. We simulated focal adhesions of varying size and distribution (to see how the accuracy is affected by it) and generated different ground truth displacements and tractions. (3/10) 
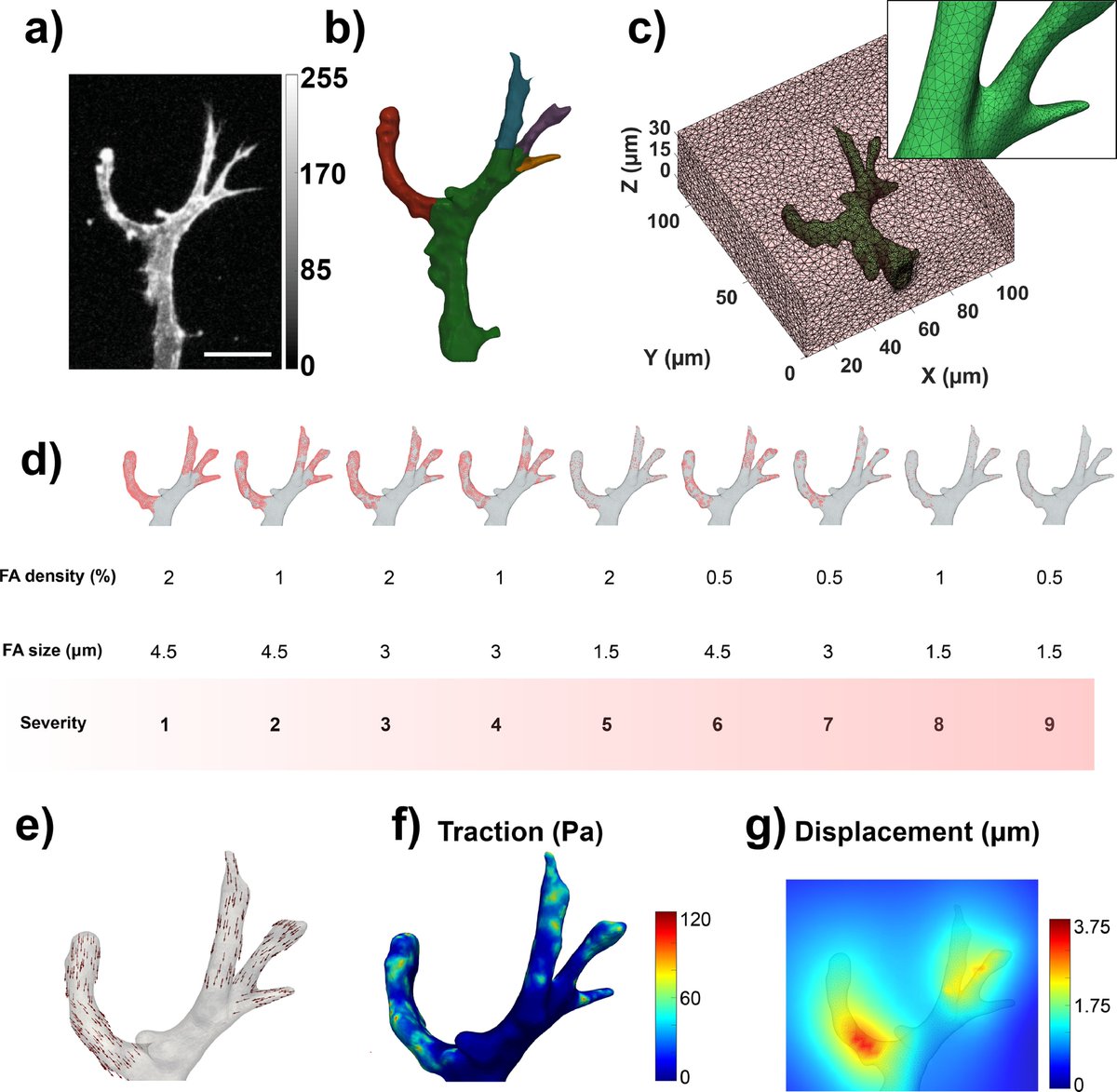
We also simulated bead images that look as similar as possible as the ones acquired with our microscope. This allows us to see the effect of bead density in our simulations as well! (4/10) 

We implemented two traction recovery methods: a forward method (which calculates the tractions directly from the measured displacements) and our novel inverse method (which imposes the mechanical equilibrium of forces in the hydrogel as a physical constraint). (5/10)
Our RESULTS show that while the displacement error is similar for both methods, our inverse method does a much better job in recovering tractions! We also see that lower and sparser focal adhesions (high severity cases) lead to higher errors. (6/10) 

We applied both methods to a real case of an angiogenic sprout invading a PEG hydrogel. Again, our inverse method retrieved more reliable traction field patterns as traction maxima were located near the sprout protrusion tips and showed clearer pulling patterns (7/10) 

Our framework can be used to test the optimal computational parameters and algorithms for a given experimental setup. This is vital to properly characterize the range of errors that you can expect from your data and avoiding obscuring physiological information. (8/10)
After such a thorough validation process, 😅, I think I can safely say that our inverse method is clearly very robust against inherent limitations of TFM (sparse and small focal adhesions or low bead densities) and that it retrieves reliable traction field patterns. (9/10)
Finally, I would also like to thank the coauthors @apek_shapeti , Janne, @AdrianRanga1 , José Antonio and Hans. I´m very proud that this is my first article, so thanks for all your contributions 💜 (10/10)
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh




